?The spinal cord links and allows communication between the brain and peripheral organs, coordinating the movement and the sensation of the body. The spinal cord formation begins around embryonic day 9 (e9) in mice. During development, the neural tube generates 11 types of neural progenitors along the dorsal ventral axis. Subsequently, these cells differentiate into neurons with different neurotransmitters which form a functional spinal cord. However, the regulatory mechanisms of this process are still poorly understood.
A recent study from collaboration between Prof. DAI Jianwu and Prof. LU Falong at the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS), as well as Prof. HUANG Jialing at the Xiamen University utilized the single-cell chromatin accessibility technique to reveal the underlying regulatory mechanism of spinal cord neural cells development.
The study entitled “Single-cell chromatin accessibility identifies enhancer networks driving gene expression during spinal cord development in mous” was published in Developmental Cell on December 19 (DOI:10.1016/j.devcel.2022.11.011).
In this study, the researhers depicted a single-cell chromatin accessibility atlas of developing mouse spinal cord and identified key cis-regulatory elements and regulators for neural cells. Then, they used eNet algorithm, a novel approach ?developed by these researchers, to construct enhancer regulatory network during mouse spinal cord development to explore how enhancers regulate cell fate decision. They found that the cell identity and disease genes ?are more likely to be regulated by complex enhancer network. Furthermore, the researchers found that the enhancer networks were organized in a hierarchical manner, in which the hub enhancer played a more important role in the function of the enhancer network. Finally, using enhancer reporter transgenic mice and enhancer knockout mice, they verified the role and mechanism of enhancer regulatory network in gene expression. They proved that absence of hub enhancers significantly affected target gene expression.
In sum, these analyses provide comprehensive resources for regulatory insight into spinal cord development, and provide compelling evidence for the functional importance of enhancer networks in regulating key developmental genes.
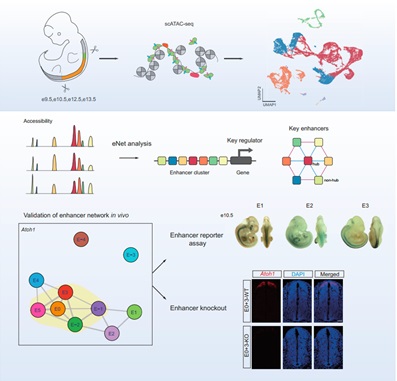
?
Enhancer networks control master regulators during spinal cord development and experimentally validate Atoh1 enhancers through in vivo perturbation as a proof of concept. (Image by IGDB)
“We focus on spinal cord injury for many years and have developped a neural regeneration collagen scaffold named NeuroRegen Scaffold. This product has been explanted in more than 100 patients in an ongoing clinical trial, and ??sufficiently restored walking after spinal cord injury caused paralysis. We are trying to improve spinal cord regeneration with the prodcut. We realize that study the developmental process of spinal cord will help us understand spinal cord regeneration. This work advances our understing of developmental process of spinal cord and have implications for improving NeuroRegen Scaffold efficeiency in the future.” Said Prof. DAI Jianwu, lead and corresponding author of the study.
“This study ?provides insights into the epigenetic regulation of spinal cord development. In addition, this work also provides potential new targets for spinal cord injury repair and treatment of related diseases.” said Prof. LU Falong, another corresponding author of the study.
This study was funded by the ?National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academy of Sciences, CAS Project for Young Scientists in Basic Research, and National Key Research and Development Program of China.
Contact:
Prof. DAI Jianwu and Prof. LU Falong
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
 ?Enhancer networks control master regulators during spinal cord development and experimentally validate Atoh1 enhancers through in vivo perturbation as a proof of concept. (Image by IGDB)“We focus on spinal cord injury for many years and have developped a neural regeneration collagen scaffold named NeuroRegen Scaffold. This product has been explanted in more than 100 patients in an ongoing clinical trial, and ??sufficiently restored walking after spinal cord injury caused paralysis. We are trying to improve spinal cord regeneration with the prodcut. We realize that study the developmental process of spinal cord will help us understand spinal cord regeneration. This work advances our understing of developmental process of spinal cord and have implications for improving NeuroRegen Scaffold efficeiency in the future.” Said Prof. DAI Jianwu, lead and corresponding author of the study.“This study ?provides insights into the epigenetic regulation of spinal cord development. In addition, this work also provides potential new targets for spinal cord injury repair and treatment of related diseases.” said Prof. LU Falong, another corresponding author of the study.This study was funded by the ?National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academy of Sciences, CAS Project for Young Scientists in Basic Research, and National Key Research and Development Program of China.Contact:Prof. DAI Jianwu and Prof. LU FalongInstitute of Genetics and Developmental Biology, Chinese Academy of Sciences
?Enhancer networks control master regulators during spinal cord development and experimentally validate Atoh1 enhancers through in vivo perturbation as a proof of concept. (Image by IGDB)“We focus on spinal cord injury for many years and have developped a neural regeneration collagen scaffold named NeuroRegen Scaffold. This product has been explanted in more than 100 patients in an ongoing clinical trial, and ??sufficiently restored walking after spinal cord injury caused paralysis. We are trying to improve spinal cord regeneration with the prodcut. We realize that study the developmental process of spinal cord will help us understand spinal cord regeneration. This work advances our understing of developmental process of spinal cord and have implications for improving NeuroRegen Scaffold efficeiency in the future.” Said Prof. DAI Jianwu, lead and corresponding author of the study.“This study ?provides insights into the epigenetic regulation of spinal cord development. In addition, this work also provides potential new targets for spinal cord injury repair and treatment of related diseases.” said Prof. LU Falong, another corresponding author of the study.This study was funded by the ?National Natural Science Foundation of China, Strategic Priority Research Program of the Chinese Academy of Sciences, CAS Project for Young Scientists in Basic Research, and National Key Research and Development Program of China.Contact:Prof. DAI Jianwu and Prof. LU FalongInstitute of Genetics and Developmental Biology, Chinese Academy of Sciences CAS
CAS
 中文
中文




.png)
