Plants encounter a wide variety of pathogens in the wild, and plant defense responses to counter this threat are tightly controlled by many positive and negative regulators to cope with attacks from various pathogens. Powdery mildew (Golovinomyces cichoracearum) is one of the biotrophic pathogens, which depend on living host cells for invasion and reproduction. Powdery mildew pathogens infect a broad range of crop species including barley, wheat, and grape, and cause large worldwide economic losses.
Arabidopsis EDR2 (ENHANCED DISEASE RESISTANCE 2) is a negative regulator of powdery mildew resistance and the edr2 mutant displays enhanced resistance to powdery mildew. Scientists in Dr. Dingzhong Tang’s Group screened for edr2 suppressors to identify components acting in the EDR2 pathway. They identified a gain-of-function mutation in SR1 (SIGNAL RESPONSIVE 1), which encodes a calmodulin-binding transcription activator. The sr1-4D gain-of-function mutation suppresses all edr2-associated phenotypes, including powdery mildew resistance, mildew-induced cell death and ethylene-induced senescence. The sr1-4D single mutant is more susceptible to a Pseudomonas syringae pv. tomato (Pto) DC3000 virulent strain and to avirulent strains carrying avrRpt2 or avrRPS4 than wild type. They found that SR1 directly binds to the promoter region of NDR1, a key component in RPS2-mediated plant immunity. Also, the ndr1 mutation suppresses the sr1-1 null allele, which shows enhanced resistance to both Pto DC3000 avrRpt2 and G. cichoracearum. They also found that SR1 regulates ethylene-induced senescence by directly binding to the EIN3 promoter region in vivo. Enhanced ethylene-induced senescence in sr1-1 is suppressed by ein3. These facts indicate that SR1 regulates plant defense responses and senescence by directly binding to the promoter regions of NDR1 and EIN3.
This work with Dr. Haozhen Nie as the first author has been online published on
Plant Physiology(
DOI:10.1104/pp.111.192310). This research was supported by grants from National Basic Research Program of China, the National Natural Science Foundation of China and the National Transgenic Program of China.
AUTHOR CONTACT:
Dingzhong Tang, Ph.D.
Institute of Genetics and Developmetnal Biology, Chinese Academy of Sciences, Beijing, China.
Figure.sr1-4Dsuppressed the edr2 phenotype of resistance to powdery mildew and ethylene induced senescence
A. Four-week-old wild type, edr2, edr2 sr1-4D plants were infected with powdery mildew G. cichoracearumUCSC1 and therepresentative leaves were removed and photographed at 8 dpi. The edr2 sr1-4D double mutant displayed a susceptible phenotype,showing visible powder and no necrosis, which was similar to wild type. Thirty plants were evaluated for each genotype.
B. Trypan blue staining to visualize plant cell death and fungal growth. Leaves were stained with trypan blue at 8 dpi. The edr2mutant displayed massive cell death and very few conidia, while edr2 sr1-4D supported wild type like conidia formation. Bar =100 μm.
C. DAB staining for H2O2 at 2 dpi. Note that edr2 accumulated more H2O2 than wild type. Bar = 100 μm.
D. Accumulation of H2O2 was quantified as described previously (Wang et al., 2011). The bars represent mean and standard deviationof intensity per area from at least 6 leaves of 3 plants for each genotype. Lower-case letters indicate a significant difference(P<0.01, one-way ANOVA). The experiments were repeated three times with similar results.
E. Quantification of the fungal growth by counting the number of conidiophores per colony at 7 dpi. The bars represent mean andstandard deviation of samples (n=25). Low-case letters indicate statistical significance (P<0.01, one-way ANOVA). The experimentwas repeated 3 times with similar results.
F. Accumulation of PR1 mRNA in edr2 was suppressed by sr1-4D. Four-week-old plants were inoculated with G. cichoracearum.Accumulation of PR1 transcripts was examined by real-time PCR and normalized to ACT8 as an internal control. The barsrepresent mean and standard deviation from three biological replicates. The asterisk indicates significant difference from wild type(P<0.01, Student’s t-test)G. Ethylene-induced senescence. Four-week-old plants were treated with 100 μl l-1ethylene for 3 days.
H. Chlorophyll content of the fourth to the sixth leaves of day 0 and day 3 after 100 μl l-1ethylene treatment. The bars represent meanand standard deviation (n=4). Statistical differences are indicated by lower-case letters (P<0.01, one-way ANOVA). The experimentwas repeated more than 3 times with the similar results.
(Image by Nie et al)
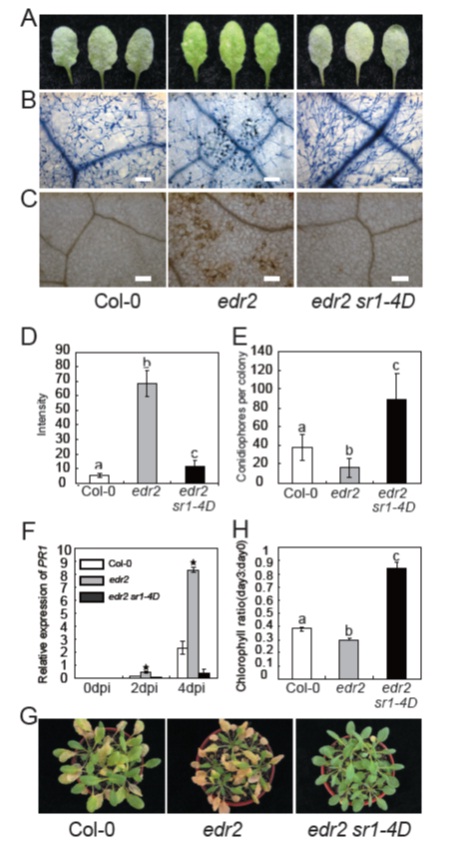 Figure.sr1-4Dsuppressed the edr2 phenotype of resistance to powdery mildew and ethylene induced senescenceA. Four-week-old wild type, edr2, edr2 sr1-4D plants were infected with powdery mildew G. cichoracearumUCSC1 and therepresentative leaves were removed and photographed at 8 dpi. The edr2 sr1-4D double mutant displayed a susceptible phenotype,showing visible powder and no necrosis, which was similar to wild type. Thirty plants were evaluated for each genotype.B. Trypan blue staining to visualize plant cell death and fungal growth. Leaves were stained with trypan blue at 8 dpi. The edr2mutant displayed massive cell death and very few conidia, while edr2 sr1-4D supported wild type like conidia formation. Bar =100 μm.C. DAB staining for H2O2 at 2 dpi. Note that edr2 accumulated more H2O2 than wild type. Bar = 100 μm.D. Accumulation of H2O2 was quantified as described previously (Wang et al., 2011). The bars represent mean and standard deviationof intensity per area from at least 6 leaves of 3 plants for each genotype. Lower-case letters indicate a significant difference(P<0.01, one-way ANOVA). The experiments were repeated three times with similar results.E. Quantification of the fungal growth by counting the number of conidiophores per colony at 7 dpi. The bars represent mean andstandard deviation of samples (n=25). Low-case letters indicate statistical significance (P<0.01, one-way ANOVA). The experimentwas repeated 3 times with similar results.F. Accumulation of PR1 mRNA in edr2 was suppressed by sr1-4D. Four-week-old plants were inoculated with G. cichoracearum.Accumulation of PR1 transcripts was examined by real-time PCR and normalized to ACT8 as an internal control. The barsrepresent mean and standard deviation from three biological replicates. The asterisk indicates significant difference from wild type(P<0.01, Student’s t-test)G. Ethylene-induced senescence. Four-week-old plants were treated with 100 μl l-1ethylene for 3 days.H. Chlorophyll content of the fourth to the sixth leaves of day 0 and day 3 after 100 μl l-1ethylene treatment. The bars represent meanand standard deviation (n=4). Statistical differences are indicated by lower-case letters (P<0.01, one-way ANOVA). The experimentwas repeated more than 3 times with the similar results.(Image by Nie et al)
Figure.sr1-4Dsuppressed the edr2 phenotype of resistance to powdery mildew and ethylene induced senescenceA. Four-week-old wild type, edr2, edr2 sr1-4D plants were infected with powdery mildew G. cichoracearumUCSC1 and therepresentative leaves were removed and photographed at 8 dpi. The edr2 sr1-4D double mutant displayed a susceptible phenotype,showing visible powder and no necrosis, which was similar to wild type. Thirty plants were evaluated for each genotype.B. Trypan blue staining to visualize plant cell death and fungal growth. Leaves were stained with trypan blue at 8 dpi. The edr2mutant displayed massive cell death and very few conidia, while edr2 sr1-4D supported wild type like conidia formation. Bar =100 μm.C. DAB staining for H2O2 at 2 dpi. Note that edr2 accumulated more H2O2 than wild type. Bar = 100 μm.D. Accumulation of H2O2 was quantified as described previously (Wang et al., 2011). The bars represent mean and standard deviationof intensity per area from at least 6 leaves of 3 plants for each genotype. Lower-case letters indicate a significant difference(P<0.01, one-way ANOVA). The experiments were repeated three times with similar results.E. Quantification of the fungal growth by counting the number of conidiophores per colony at 7 dpi. The bars represent mean andstandard deviation of samples (n=25). Low-case letters indicate statistical significance (P<0.01, one-way ANOVA). The experimentwas repeated 3 times with similar results.F. Accumulation of PR1 mRNA in edr2 was suppressed by sr1-4D. Four-week-old plants were inoculated with G. cichoracearum.Accumulation of PR1 transcripts was examined by real-time PCR and normalized to ACT8 as an internal control. The barsrepresent mean and standard deviation from three biological replicates. The asterisk indicates significant difference from wild type(P<0.01, Student’s t-test)G. Ethylene-induced senescence. Four-week-old plants were treated with 100 μl l-1ethylene for 3 days.H. Chlorophyll content of the fourth to the sixth leaves of day 0 and day 3 after 100 μl l-1ethylene treatment. The bars represent meanand standard deviation (n=4). Statistical differences are indicated by lower-case letters (P<0.01, one-way ANOVA). The experimentwas repeated more than 3 times with the similar results.(Image by Nie et al) CAS
CAS
 中文
中文




.png)
