The crucifer Thellungiella salsuginea, a close relative of Arabidopsis, originally classified as Thellungiella halophila, is a halophyte with relative high resistance to cold, drought and oxidative stresses. T. salsuginea is exemplary by its short life cycle, self-fertility, relative small genome and being genetically transformable. These characters make the species an excellent extremophile model for unraveling factors constituting to abiotic stress tolerance.
Scientists from Dr. Qi Xie, Dr. Xiu-Jie Wang, Dr. Shou-Yi Chen and Chengcai Chu’s groups of Institute of Genetics and Developmental Biology, the Chinese Academy of Sciences and other co-workers from BGI-Shenzhen, Purdue University and University of Illinois at Urbana-Champaign have carried out genome sequencing of T. salsuginea using Illumina GA II technology, and present here the draft sequence genome based on ~134-fold coverage of the estimated 260Mb genome.
Reaserchers assembled 233.7Mb scaffold sequences and anchored 80% of them in 7 chromosomes through a hybrid and hierarchical assembly approach. A total of 28,457 protein coding regions were predicted in the sequenced T. salsuginea genome. Among the predicted genes, 92.55% can be functional classified and 6.45% probably represent the specific gene groups of T. salsuginea. The T. salsuginea genome is approximately twice that of A. thaliana in size. A substantial part of that difference is due to a proliferation of transposable elements (TEs) representing ~52% of the genome, which is about four fold of that in A. thaliana. Phylogenetic analyses indicate a time of divergence of 7-12 MYA between T. salsuginea and A. thaliana, after which there is no whole genome duplication events. Term “response to stimulus” was identified among the significantly differed GO categories between T. salsuginea and A. thaliana, with more genes classified into this category in the T. salsuginea genome.
Comparative genomics and experimental analyses revealed that genes related to ABA signaling, cation transport and wax production prominent in T. salsuginea may contribute to its success in stressful environments. This genome provides resources and previously unavailable evidence about the nature of defense mechanisms constituting the genetic basis underlying plant abiotic stress tolerance, which will largely promote the studies on plant salinity resistance in the future.
The above work has been published online in Proceedings of the National Academy of Sciences of the USA (PNAS). Hua-Jun Wu, Zhonghui Zhang, Junyi Wang, Dong-Ha Oh, Maheshi Dassanayake, Binghang Liu and Quanfei Huang contribute equally as the first authors. This study is supported by grants from National Science Foundation of China, National Basic Research Program of China, the 973 project, State Key Laboratory of Plant Genomics of China.
AUTHOR CONTACT:
(Image by Dong-Ha Oh et al.)
Figure 1. The assembled genome of T. salsuginea.
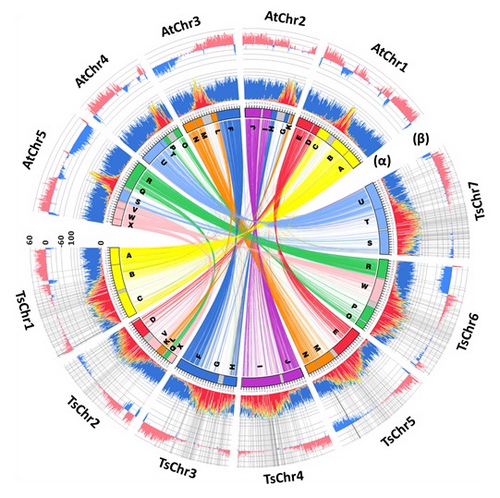 (Image by Dong-Ha Oh et al.)Figure 1. The assembled genome of T. salsuginea.
(Image by Dong-Ha Oh et al.)Figure 1. The assembled genome of T. salsuginea. CAS
CAS
 中文
中文




.png)
