AtDOF4.2-overexpressing plants exhibit an increased branching phenotype (Figure 1), and the mutation of Thr-Met-Asp motif in AtDOF4.2 significantly reduces the branching in transgenic plants. AtDOF4.2 may achieve this function through the upregulation of three branching-related genes, AtSTM, AtTFL1 and AtCYP83B1. The seeds of the AtDOF4.2-overexpressing plant show collapse-like morphology in epidermal cells of the seed coat (Figure 2). Mucilage contents and the concentration and composition of mucilage monosaccharides are significantly changed in the seed coat of transgenic plants. AtDOF4.2 may exert its effects on the seed epidermis through the direct binding and activation of the cell wall loosening-related gene AtEXPA9. The dof4.2 mutant did not exhibit changes in branching or its seed coat; however, the silique length and seed yield were increased. AtDOF4.4, which is a close homolog of AtDOF4.2, also promotes shoot branching and affects silique size and seed yield. Manipulation of these genes should have a practical use in the improvement of agronomic traits in important crops.
This work has been published online in Biochemical Journal (doi:10.1042/BJ20110060) on Oct 24, 2012, with Hong-Feng Zou and Yu-Qin Zhang as the co-first authors. Prof Yi-Hua Zhou’s lab also participated in the work. This study is supported by 973 project, Transgenic Research project and National Natural Science Foundation of China.
Figure 1 Overexpression of AtDOF4.2 promotes shoot branching in transgenic plants. (a) AtDOF4.2 expression in various transgenic lines as revealed by Northern analysis. The rRNAs are shown as loading controls. (b) Identification of the AtDOF4.2 T-DNA insertion mutant dof4.2. No AtDOF4.2 expression was found in dof4.2 (right panel). (c) Shoot phenotype in different plants. AtDOF4.2-overexpression lines 4.2-4 and 4.2-8 show a bushy phenotype. (d) Number of primary rosette branches (RI). (e) Number of secondary rosette branches (RII). (f) Number of primary cauline branches (CI). (g) Number of secondary cauline branches (CII). (h) Plant height of different plant lines. (i) First internode length of different lines. From (d) to (i), ‘**’ indicate highly significant differences compared to Col-0 (P <0.01). Bars indicate SD (n=32).
Figure 2 Seed coat phenotypes of various plants. Mutant dof4.2 and lines overexpressing AtDOF4.2 (4.2-4 and 4.2-8) or mutated AtDOF4.2 (4.2-m-16 and 4.2-m-17) were compared with Col-0. (a) Scanning electron micrograph of seed coat. Bars represent 40 μm. (b) Scanning electron micrograph of epidermal cells in seed coat. Bars represent 10 μm. (c) Ruthenium red staining of seeds. Seeds of transgenic lines 4.2-4 and 4.2-8 showed different staining patterns from those of line 4.2-m-17 and Col-0. Bars represent 500 μm. (d) Percentage of stained seeds in various plants. Bars indicate SD (n=3). ‘**’ indicate significant difference from Col-0 (P<0.01).
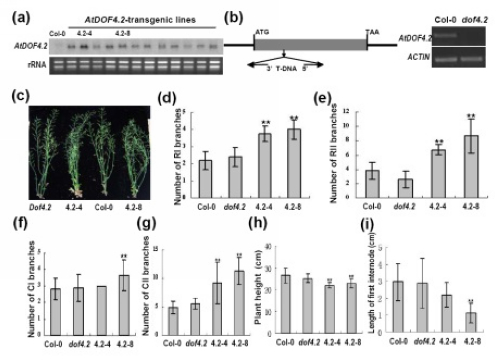 Figure 1 Overexpression of AtDOF4.2 promotes shoot branching in transgenic plants. (a) AtDOF4.2 expression in various transgenic lines as revealed by Northern analysis. The rRNAs are shown as loading controls. (b) Identification of the AtDOF4.2 T-DNA insertion mutant dof4.2. No AtDOF4.2 expression was found in dof4.2 (right panel). (c) Shoot phenotype in different plants. AtDOF4.2-overexpression lines 4.2-4 and 4.2-8 show a bushy phenotype. (d) Number of primary rosette branches (RI). (e) Number of secondary rosette branches (RII). (f) Number of primary cauline branches (CI). (g) Number of secondary cauline branches (CII). (h) Plant height of different plant lines. (i) First internode length of different lines. From (d) to (i), ‘**’ indicate highly significant differences compared to Col-0 (P <0.01). Bars indicate SD (n=32).Figure 2 Seed coat phenotypes of various plants. Mutant dof4.2 and lines overexpressing AtDOF4.2 (4.2-4 and 4.2-8) or mutated AtDOF4.2 (4.2-m-16 and 4.2-m-17) were compared with Col-0. (a) Scanning electron micrograph of seed coat. Bars represent 40 μm. (b) Scanning electron micrograph of epidermal cells in seed coat. Bars represent 10 μm. (c) Ruthenium red staining of seeds. Seeds of transgenic lines 4.2-4 and 4.2-8 showed different staining patterns from those of line 4.2-m-17 and Col-0. Bars represent 500 μm. (d) Percentage of stained seeds in various plants. Bars indicate SD (n=3). ‘**’ indicate significant difference from Col-0 (P<0.01).
Figure 1 Overexpression of AtDOF4.2 promotes shoot branching in transgenic plants. (a) AtDOF4.2 expression in various transgenic lines as revealed by Northern analysis. The rRNAs are shown as loading controls. (b) Identification of the AtDOF4.2 T-DNA insertion mutant dof4.2. No AtDOF4.2 expression was found in dof4.2 (right panel). (c) Shoot phenotype in different plants. AtDOF4.2-overexpression lines 4.2-4 and 4.2-8 show a bushy phenotype. (d) Number of primary rosette branches (RI). (e) Number of secondary rosette branches (RII). (f) Number of primary cauline branches (CI). (g) Number of secondary cauline branches (CII). (h) Plant height of different plant lines. (i) First internode length of different lines. From (d) to (i), ‘**’ indicate highly significant differences compared to Col-0 (P <0.01). Bars indicate SD (n=32).Figure 2 Seed coat phenotypes of various plants. Mutant dof4.2 and lines overexpressing AtDOF4.2 (4.2-4 and 4.2-8) or mutated AtDOF4.2 (4.2-m-16 and 4.2-m-17) were compared with Col-0. (a) Scanning electron micrograph of seed coat. Bars represent 40 μm. (b) Scanning electron micrograph of epidermal cells in seed coat. Bars represent 10 μm. (c) Ruthenium red staining of seeds. Seeds of transgenic lines 4.2-4 and 4.2-8 showed different staining patterns from those of line 4.2-m-17 and Col-0. Bars represent 500 μm. (d) Percentage of stained seeds in various plants. Bars indicate SD (n=3). ‘**’ indicate significant difference from Col-0 (P<0.01). CAS
CAS
 中文
中文




.png)
