Abscisic acid (ABA) is essential for plant adaptation to abiotic stresses. In the past several years, great progress has been made in understanding of ABA signal perception and the signaling transduction pathway with discovery of the ABA receptors (PYR/PYL)-Group A protein type 2C phosphatases (PP2C)-protein kinases (SnRK2) core regulatory module. Apparently, the PYR/PYL-PP2C-SnRK2 core module mainly regulates the downstream effectors and plant response to ABA at the post-translational level. The reports on ABA regulation at post-transcriptional level remain scarce.
Alternative splicing (AS) of pre-mRNAs is emerging as a key step that regulates transcript levels and the subsequent transcript isoforms of genes, giving rise to proteins that differ in subcellular localization, stability, and function. More than 61% of the intron-containing Arabidopsis genes undergo AS under normal conditions. The percentage of genes showing AS will dramatically increase when plants are subjected to abiotic stresses. Thus, AS plays a fundamental role in plant development and stress adaptation. However, the role of AS in regulating ABA signaling and plant stress adaptations remains elusive.
PP2Cs are central components in the ABA core regulatory module, and have long been considered as the key negative regulators of the ABA signaling. Hypersensitive to ABA1 (HAB1) is a member of PP2Cs and negatively regulates ABA signaling by inhibiting autophosphorylation of the protein kinase SnRK2s. However, it seems that HAB1 gene plays a more subtle role in regulation of the ABA signaling.
Most recently, scientists in Dr. LI Xia’s group from the Institute of Genetics and Development Biology, the Chinese Academy of Sciences, found that the HAB1 pre-mRNA undergoes AS to produce two splice variants, which encode HAB1.1 and HAB1.2. HAB1.1 is the previous identified HAB1 with the phosphotase catalytic domain, whereas HAB1.2 is a truncated protein lacking the catalytic domain. Intriguingly, HAB1.1 and HAB1.2 play opposing roles in ABA-mediated seed germination and ABA-mediated post-germination developmental arrest. HAB1.2 is predominately formed in the presence of ABA and prevents seed germination and post-germinative growth. HAB1.2 is capable to interact with OST1 like HAB1.1, but cannot inhibit OST1 kinase activity; thus, it functions as a positive regulator of ABA signaling.
Furthermore, the scientists identified a RNA recognition motif (RRM)-containing protein, RBM25, as a potential regulator of HAB1 alternative splicing and molecular diversity. Thus, these findings reveal a novel mechanism at the post-transcriptional level for turning ABA signaling on and off and for plant adaptation to abiotic stress.
This research was supported National Science Foundation (31230050 and 31200220), the Ministry of Agriculture of the People’s Republic of China (2013ZX08009-02-03), National Program on Key Basic Research Project (2012CB114300).
AUTHOR CONTACT:
LI Xia, Group Leader
The State Key Laboratory of Plant Cell & Chromosome Engineering, Center for Agricultural Research Resources, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, 286 Huaizhong Road, Shijiazhuang, Hebei 050021, P.R. China; State Key Laboratory of Agricultural Microbiology, College of Plant Science and Technology, Huazhong Agricultural University, Wuhan 430070, P.R. China.
Tel: 86-311-85871744
Figure. A model of RBM25-mediated HAB1alternative splicing by which HAB1 splice variants control ABA signalling. (Image by IGDB)
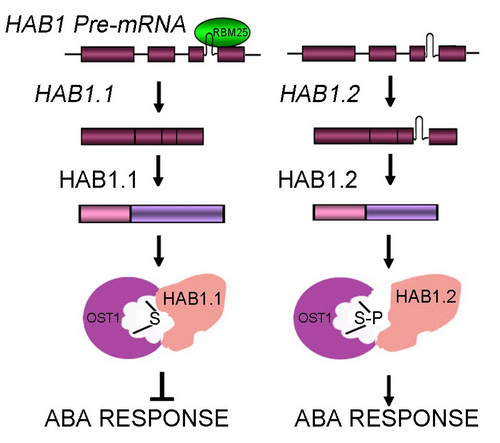 Figure. A model of RBM25-mediated HAB1alternative splicing by which HAB1 splice variants control ABA signalling. (Image by IGDB)
Figure. A model of RBM25-mediated HAB1alternative splicing by which HAB1 splice variants control ABA signalling. (Image by IGDB) CAS
CAS
 中文
中文




.png)
