Long noncoding RNAs (lncRNAs) are found widespread in many organisms. However, their biological functions remain largely unknown, particularly in plants. Leaves are the main photosynthetic organs and maintaining the leaf blade flattening is of great importance in plants. However, the molecular mechanism involved in the leaf blade flattening is largely unknown
The group of Prof. ZHU Lihuang at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences (CAS), and the group of Prof. HU Songnian at the Beijing Genomics Institute, CAS, recently reported that a cis-natural lncRNA could affect the rice leaf morphology.
This lncRNA locus was named as TWISTED LEAF (TL) according to its RNAi knock-down phenotype and was transcribed from the opposite strand of a protein coding gene, OsMYB60. The TL and OsMYB60 transcripts both accumulate in mostly the same tissues, suggesting that they may function together and that TL might regulate OsMYB60.
Then, the scientists found that expression of OsMYB60 was increased in the TL RNAi knock-down transgenic lines and that over-expression of OsMYB60 could produce a similar twisted leaf phenotype to the TL RNAi-lines. Considering that some lncRNAs function through chromatin modification approach, the scientists then assayed the chromatin at OsMYB60 locus in the TL-RNAi transgenic lines using ChIP-PCR. The repressive chromatin marks were enriched at OsMYB60 locus in wild type tissues while active marks were enriched in the TL-RNAi lines, suggesting TL functioned to repress OsMYB60 through regulation of chromatin modifications.
They further discovered that a zinc finger protein, OsZFP7, could directly interact with OsMYB60. Notably, the OsZFP7 transcript accumulation was also higher in both the OsMYB60 overexpressing lines and the TL-RNAi lines, and over expression of OsZFP7 also produced the twisted leaf phenotype similar to those of the OsMYB60 overexpression and TL-RNAi lines. These results suggest that OsMYB60 functions with OsZFP7 in a complex to regulate leaf blade shape in rice.
Overall, the results demonstrate that expression of OsMYB60 is epigenetically regulated by the lncRNA TL and both function together to maintain leaf blade flattening in rice.
The relevant work entitled “
A novel antisense long noncoding RNA, TWISTED LEAF, maintains leaf blade flattening by regulating its associated sense R2R3-MYB gene in rice” has been online published in
New Phytologist on Feb 7, 2018 (
DOI:10.1111/nph.15023), with Dr. LIU Xue and Dr. LI Dayong as the first authors.
This work was supported by the National Key Research and Development Program of China (2016YFD0100904), the Major Project of New Varieties of Genetically Modified Organism of China (2016ZX08009-001), and the State Key Laboratory of Plant Genomics, China (2017A0407-19).
Figure 1. OsMYB60 is epigenetically regulated by the antisense lncRNA TWISTED LEAF (TL) to mantain leaf blade flattening in rice. (Image by IGDB)
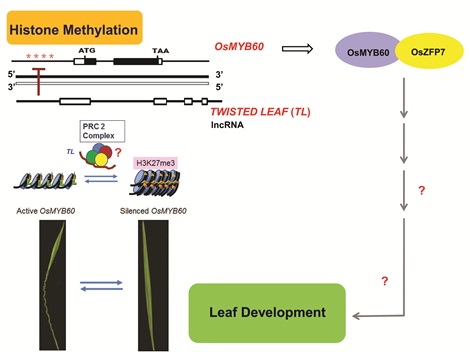 Figure 1. OsMYB60 is epigenetically regulated by the antisense lncRNA TWISTED LEAF (TL) to mantain leaf blade flattening in rice. (Image by IGDB)CONTACT:
Figure 1. OsMYB60 is epigenetically regulated by the antisense lncRNA TWISTED LEAF (TL) to mantain leaf blade flattening in rice. (Image by IGDB)CONTACT: CAS
CAS
 中文
中文




.png)
