The CRISPR-Cas9 system, which is derived from a bacterial immune system, has been used as a universal genome editing tool for various species including plants.
For vertebrates, in which the Cas9 (mRNA or protein) and sgRNAs could be simultaneously injected into zygotic cells, the dose can be controlled to enhance the editing efficiency. However, for most crop plants, transformation mostly depends on Agrobacterium mediated transformation. The dose of Cas9 in receptor cells depends on the regulatory elements such as the promoter used.
Previously in plants such as maize, the genome editing efficiency is relatively low when the vectors are delivered by Agrobacterium. The low efficiency is an impediment for application of CRISPR-Cas9, especially when conducting a high throughput mutant screening project.
Recently, researchers from HAN Fangpu's group in the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences (CAS), reported the dmc1 promoter for Cas9 that could enhance the editing efficiency in maize.
In the study, the promoters of several meiosis-specific genes were tested for expressing Cas9. The authors were surprised to found that one of them, the dmc1 promoter, could generate highly-efficient genome editing in calli and T0 seedlings. About 60%-70% T0 seedlings were homozygous or bi-allelic mutant plants, with the rest being heterozygotes or chimeras.
The genotyping results also revealed that seedlings regenerated from the same callus have different mutant alleles.
No off-target mutation was detected in the predicted sites in T0 seedlings by genome-wide high throughput sequencing. As the DMC1 is evolutionarily conserved, this promoter could possibly be used for other plants.
This work entitled "High efficiency genome editing by using
dmc1 promoter-controlled CRISPR/Cas9 system in maize" was online published in
Plant Biotechnology Journal (
DOI:10.1111/pbi.12920 ).
This work is supported by grants from the National Key Research and Development Program of China, the State Key Program of National Natural Science Foundation of China and the Ministry of Science and Technology (MOST) Project.
Figure 1. Targeted editing of zb7 gene by DPC CRISPR-Cas9 system. A. The gene structure, target site of zb7 and the construct of DPC CRISPR/Cas9; B. The phenotype of T0 plant with zb7 edited; C. Genotyping results of partial T0 mutant plants. (Image by IGDB)
Contact:
Dr. HAN Fangpu
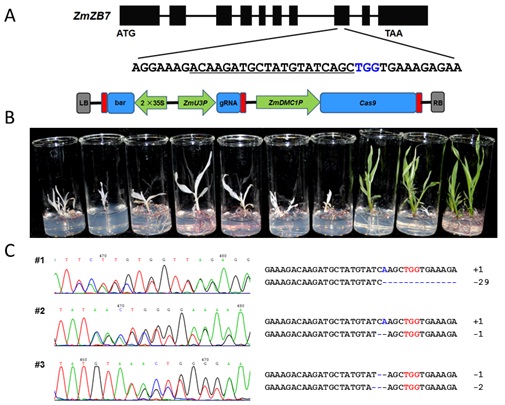 Figure 1. Targeted editing of zb7 gene by DPC CRISPR-Cas9 system. A. The gene structure, target site of zb7 and the construct of DPC CRISPR/Cas9; B. The phenotype of T0 plant with zb7 edited; C. Genotyping results of partial T0 mutant plants. (Image by IGDB)Contact:Dr. HAN FangpuEmail: fphan@genetics.ac.cn
Figure 1. Targeted editing of zb7 gene by DPC CRISPR-Cas9 system. A. The gene structure, target site of zb7 and the construct of DPC CRISPR/Cas9; B. The phenotype of T0 plant with zb7 edited; C. Genotyping results of partial T0 mutant plants. (Image by IGDB)Contact:Dr. HAN FangpuEmail: fphan@genetics.ac.cn CAS
CAS
 中文
中文




.png)
