Many small regulatory elements, including miRNAs, miRNA binding sites, and cis-acting elements, comprise only about 5~24 nucleotides and play important roles in regulating gene expression, transcription and translation, and protein structure, and thus are promising targets for gene function studies and crop improvement.
The CRISPR–Cas9 system has been widely applied for genome engineering. In this system, a sgRNA-guided Cas9 nuclease generates chromosomal double-strand breaks (DSBs), which are mainly repaired by nonhomologous end joining (NHEJ), resulting in frequent short insertions and deletions (indels) of 1~3 bp. However, the heterogeneity of these small indels makes it technically challenging to disrupt these regulatory DNAs. Thus, the development of precise, predictable multi-nucleotide deletion system is of great significance to gene function analysis and application of these regulatory DNAs.
The research team led by Prof. GAO Caixia of the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences focuses on developing novel technologies to achieve efficient and specific genome engineering. Based on the mechanism of cytidine deamination and base excision repair (BER), the researchers developed a series of APOBEC-Cas9 fusion-induced deletion systems (AFIDs) that combine Cas9 with human APOBEC3A (A3A), uracil DNA-glucosidase (UDG) and AP lyase and successfully induced novel precise, predictable multi-nucleotide deletions in the rice and wheat genomes.
Their results showed that AFID-3 produced a variety of predictable deletions extending from the 5′-deaminated Cs to the Cas9 cleavage sites, with the average predicted proportions over 30%. The researchers further screened the deamination activity of different cytosine deaminases in rice protoplasts, and found that the truncated APOBEC3B (A3Bctd) that displayed not only a higher base-editing efficiency but also a narrower window than other deaminases. They therefore replaced A3A in AFID-3 with A3Bctd, generating eAFID-3. eAFID-3 produced more uniform deletions from the preferred TC motifs to double-strand breaks, 1.52-fold higher than that of AFID-3. Moreover, the researchers used the AFID system to target the effector-binding element of OsSWEET14 in rice, and found that the predictable deletion mutants conferred enhanced resistance to rice bacterial blight.
AFID systems are superior to other current tools for generating predictable multi-nucleotide targeted deletions within the protospacer, and thus promise to provide robust deletion tools for basic research and genetic improvement.
The research was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences, the National Transgenic Science and Technology Program, the National Natural Science Foundation of China.
Figure: AFID systems induce precise and predictable multi-nucleotide deletions in plants (Image by IGDB). (a) Schematic representation of the AFID system. (b) Structures of AFIDs 1–3 and eAFID-3. (c) The proportions of predictable deletions generated by Cas9 and AFID-3. (d) Comparison of predictable deletion types between AFID-3 and eAFID-3. (e) Differences in bacterial blight resistance between 1~2 bp indel mutants and predicted deletion mutants induced by AFID-3 in the effector-binding elements of OsSWEET14.
Contact:
QI Lei
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: lqi@genetics.ac.cn
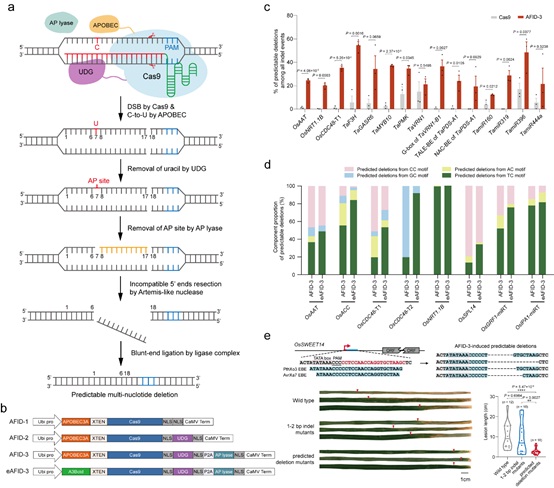 Figure: AFID systems induce precise and predictable multi-nucleotide deletions in plants (Image by IGDB). (a) Schematic representation of the AFID system. (b) Structures of AFIDs 1–3 and eAFID-3. (c) The proportions of predictable deletions generated by Cas9 and AFID-3. (d) Comparison of predictable deletion types between AFID-3 and eAFID-3. (e) Differences in bacterial blight resistance between 1~2 bp indel mutants and predicted deletion mutants induced by AFID-3 in the effector-binding elements of OsSWEET14.Contact:QI LeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: lqi@genetics.ac.cn
Figure: AFID systems induce precise and predictable multi-nucleotide deletions in plants (Image by IGDB). (a) Schematic representation of the AFID system. (b) Structures of AFIDs 1–3 and eAFID-3. (c) The proportions of predictable deletions generated by Cas9 and AFID-3. (d) Comparison of predictable deletion types between AFID-3 and eAFID-3. (e) Differences in bacterial blight resistance between 1~2 bp indel mutants and predicted deletion mutants induced by AFID-3 in the effector-binding elements of OsSWEET14.Contact:QI LeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: lqi@genetics.ac.cn CAS
CAS
 中文
中文




.png)
