Rice provides the staple food for more than half of the world’s population and is also a model plant for monocots. Cell differentiation of a multicellular organism relies on the gene expression diversity among cells, whose high-throughput exploration at the cellular resolution is recently enabled by the single-cell RNA sequencing (scRNA-seq) technology.
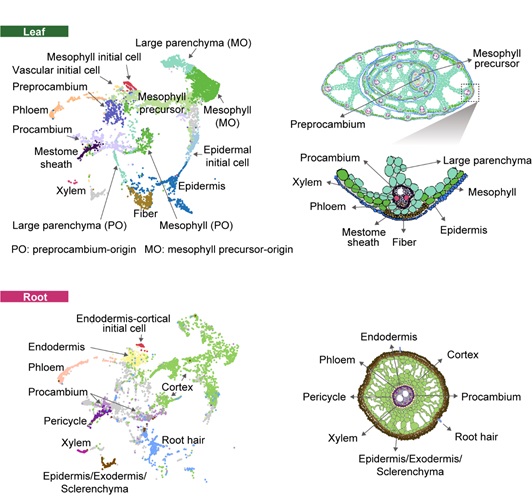
Prof. QIAN Wenfeng’s group from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences applied single-cell RNA sequencing to both shoot and root of rice seedlings and gauged the transcriptome of a total of 237,431 individual cells. They identified the cell type of each cell using the reported cell type marker genes and in situ hybridization and constructed the single-cell transcription landscapes of rice seedling. They observed that common transcriptome features were often shared between leaves and roots in the same tissue layer (for example, the vascular tissues, mesophyll cells and root cortex cells), except for the epidermis, which may be related to the difference in the external environment of the epidermal tissues.
The authors also compared the differences in single-cell transcriptome atlas of rice seedlings under multiple growth conditions and found that abiotic stress stimuli altered gene expression largely in a cell-type-specific manner. However, for a given cell type, different stresses often triggered transcriptional regulation of roughly the same set of genes. Besides, they detected proportional changes in cell populations in response to abiotic stress and investigated the underlying molecular mechanisms through single-cell reconstruction of the developmental trajectory. They observed that genes with different functions dynamically responded upon abiotic stress at various stages of mesophyll cell development, which provides clues for understanding the variation in cell differentiation upon abiotic stress.
This study represents a benchmark-setting data resource of single-cell transcriptome atlas for rice seedlings and an illustration of exploiting such resources to drive discoveries in plant biology.
This study, entitled “Single-cell transcriptome atlas of the leaf and root of rice seedlings”, has been published in
Journal of Genetics and Genomics (
DOI:10.1016/j.jgg.2021.06.001).
This study was supported by grants from the National Natural Science Foundation of China and the State Key Laboratory of Plant Genomics.
Visualization of the leaf and root cell types on the UMAP graph and the corresponding anatomic schematic. (Image by IGDB)
Contact:
Dr. QIAN Wenfeng
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
 Visualization of the leaf and root cell types on the UMAP graph and the corresponding anatomic schematic. (Image by IGDB)Contact:Dr. QIAN WenfengInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: wfqian@genetics.ac.cn
Visualization of the leaf and root cell types on the UMAP graph and the corresponding anatomic schematic. (Image by IGDB)Contact:Dr. QIAN WenfengInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: wfqian@genetics.ac.cn CAS
CAS
 中文
中文




.png)
