Balance between protein synthesis and degradation is critical for cellular homeostasis. The ubiquitin-26S proteasome system (UPS) is one of essential pathways for protein degradation in eukaryotes, and specifically ubiquitinates and degrades short-lived, misfolded and damaged proteins in a proteasome-dependent manner. Protein ubiquitination is catalyzed sequentially by ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin ligase (E3). The in vitro ubiquitination assay is a powerful tool to investigate relationships between E3 ligases and target proteins. However, the in vitro ubiquitination assay system for multisubunit E3 ligases remain difficult to achieve, especially in plants, mainly due to the complexity of composition and the difficulty in achieving active components of multisubunit E3 ligases with high purity.
The research group led by Prof. LI Jiayang from the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences, has provided a novel platform to reconstitute multisubunit SCF E3 ligases in vitro using an engineered SKP1-Cullin1-RBX1 (eSCR) protein. The study entitled “An engineered platform for reconstituting functional multisubunit SCF E3 ligase in vitro” was published online as a resource article in Molecular Plants on June 24, 2022 (DOI: 10.1016/j.molp.2022.06.011).
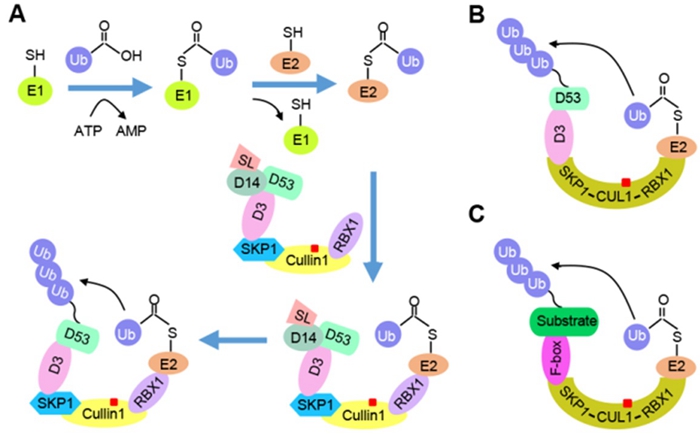
In this study, researchers took the rice SCFD3 E3 ligase from the strigolactone (SL) signaling pathway as an example, identified components of the SCFD3 E3 ligase by LC-MS/MS assay and purified active D3, Cullin1, SKP1, and RBX1 proteins from eukaryotic and prokaryotic expression systems. OsUBC14 was further identified as an OsE2 that coordinates with the SCFD3 E3 ligase in vitro. The reconstituted SCFD3 E3 ligase efficiently catalyzed the ubiquitination of its substrate protein D53 and more importantly, D53 ubiquitination was enhanced with the addition of SL receptor D14 and SL analogue rac-GR24. These results demonstrated that the functional multisubunit SCFD3 E3 ligase was reconstituted in vitro and could serve as an effective platform to identify and study essential components, active chemicals and regulatory mechanisms of SL perception.
To broaden the application of the reconstitution system, researchers further engineered SKP1, Cullin1 and RBX1 into a fusion protein SKP1-Cullin1-RBX1 (eSCR), and reconstituted theeSCFD3 E3 ligase. Importantly, the eSCFD3 E3 ligase catalyzed D53 ubiquitination as efficiently as the wild-type SCFD3 E3 ligase. The F-box D3 was further changed into other F-box proteins, including the rice GID2 that is required for gibberellin signaling, the human Fbxl18 that is essential for apoptosis and cell cycle progression, and the human CDC4 that is a critical tumor suppressor. Using the fusion protein eSCR, functional eSCFGID2 E3, eSCFFbxl18 E3, and eSCFCDC4 E3 ligases were reconstituted in vitro, and eSCFCDC4 E3 could catalyze ubiquitination of the target protein HsSic1.
In summary, the researchers provided a novel platform using the fusion protein eSCR to reconstitute different SCF E3 ligases with various F-box proteins. The powerful platform provides a practical method to make point-to-point identifications between E3s and substrates, shedding new light on mechanism research of multisubunit SCF E3 ligases in eukaryotes and providing essential parameters to study other types of multisubunit E3 ligases.
This research was supported by National Natural Science Foundation of China and Chinese Academy of Sciences.
Schematic Graphics of Platforms for Reconstituting Multisubunit SCF E3 Ligase in vitro. (Image by IGDB)
(A) Canonical SCF ubiquitin system. (B) Simplified eSCR reconstitution platform. (C) General eSCR reconstitution platform.
Contact:
Dr. LI Jiayang
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
 Schematic Graphics of Platforms for Reconstituting Multisubunit SCF E3 Ligase in vitro. (Image by IGDB)(A) Canonical SCF ubiquitin system. (B) Simplified eSCR reconstitution platform. (C) General eSCR reconstitution platform.Contact:Dr. LI JiayangInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: lqi@genetics.ac.cn
Schematic Graphics of Platforms for Reconstituting Multisubunit SCF E3 Ligase in vitro. (Image by IGDB)(A) Canonical SCF ubiquitin system. (B) Simplified eSCR reconstitution platform. (C) General eSCR reconstitution platform.Contact:Dr. LI JiayangInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: lqi@genetics.ac.cn CAS
CAS
 中文
中文




.png)
