Researchers from the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS) identified the first monocot plant viral resistance gene encoding a nucleotide-binding, leucine-rich repeat immune receptor (NLR) protein from a wild grass Brachypodium to improve the cereal crops (wheat and barley) resistance to Barley stripe mosaic virus (BSMV). The results were published online in the New Phytologist (doi.org/10.1111/nph.18457).
Viral diseases seriously threaten the crop productivity. Although a number of NLR proteins conferring resistance to specific viruses have been identified in dicot plants, NLR protein involved in viral resistance has not been found ever in monocot plants, including the most important cereal crops wheat, rice, maize, barley, oat, etc. BSMV is a positive-strand tripartite RNA virus that infects barley (Hordeum vulgare L.), wheat (Triticum aestivum L.) and Oat (Avena sativa L.), where it causes severe yield losses. During the past two decades, major progress has been made towards the understanding of infection processes of BSMV, however, studies of the incompatible interactions during grass-virus interactions are lagging behind. Therefore, identification and mining of novel BSMV resistance genes would provide a basis for preventing diseases caused by BSMV.
Brachypodium distachyon, a member of the Poaceae subfamily Pooideae, has emerged as a model species for the study of cool season cereal crops (barley, wheat, oats and rye). This small plant is easy to cultivate, has a small genome, a short life cycle, is self-fertile and has a large amount of genetic variation.
In this study, researchers cloned the first BSMV resistance gene barley stripe resistance 1 (BSR1) that encodes a typical CC-NBS-LRR (NLR) protein from B. distachyon inbred line Bd3-1 by using map-based cloning. They found the BSR1 gene from wild grass Brachypodium can be used to improve BSMV resistance in wheat and barley. Sequence variation analysis revealed that BSR1 was only present in a few B. distachyon accessions collected from Turkey-Iraq and B. hybridum accessions collected from Israel. The susceptible gene bsr1 is quite interesting with a unique region (UR) insertion at the C-terminal, resulting loss of hypersensitive response (HR) typically induced by the BSMV virus Triple Gene Block 1 (TGB1) movement protein. Using biological assays, confocal microscopy, mutational analyses, and immunoprecipitation, the authors convincingly show that the TGB1 amino acids 390/392 are key to the interaction with BSR1. In the Brachypodium BSR1 protein, the authors have identified two amino acids G196/K197 at the P-loop motif being crucial for the BSR1-TGB1 interaction. The BSR1-TGB1 interactions also occur in barley, wheat and N. benthamiana, showing the importance of using model plant Brachypodium to find novel genes for cereal crop improvement and to dissect the innate immunity mystery of monocot plant against virus.
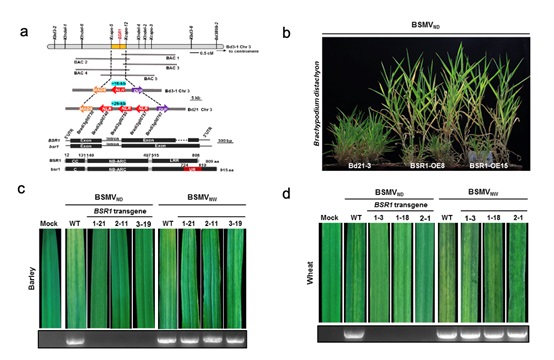
Figure: The CC‐NB‐LRR protein BSR1 from Brachypodium confers resistance to Barley stripe mosaic virus in gramineous plants (Image by IGDB)
Contact:
Dr. LIU Zhiyong
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
 Figure: The CC‐NB‐LRR protein BSR1 from Brachypodium confers resistance to Barley stripe mosaic virus in gramineous plants (Image by IGDB)Contact:Dr. LIU ZhiyongInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: zyliu@genetics.ac.cn
Figure: The CC‐NB‐LRR protein BSR1 from Brachypodium confers resistance to Barley stripe mosaic virus in gramineous plants (Image by IGDB)Contact:Dr. LIU ZhiyongInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: zyliu@genetics.ac.cn CAS
CAS
 中文
中文




.png)
