Translation, a process that expresses the genetic information from messenger RNA (mRNA) to protein, is vital to maintain cellular protein homeostasis. To synthesize functional proteins and avoid toxic translation products, the recognition of the correct initiation codon (AUG) by the small ribosomal subunit is crucial.
In the 1980s, Marilyn Kozak proposed the famous “first-AUG rule”, which asserts that the most upstream (i.e., 5′) AUG triplet is preferentially used as the primary initiation codon; insertion of an additional AUG triplet upstream (uAUG) of the annotated AUG triplet (aAUG) can significantly reduce translation initiation at the aAUG. Based on these observations, the “strictly unidirectional scanning model” was raised and commonly introduced in molecular biology textbooks. This model asserts that during eukaryotic translation initiation, the small ribosomal subunit scans exclusively in the 5′–3′ direction; if “leaky scanning” occurs, translation will initiate from the following-up AUG, and so on.
A recent study delivered by Prof. QIAN Wenfeng’s group at the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences (CAS) challenges the first-AUG rule and the strictly unidirectional scanning model. Their data revealed the small ribosomal subunits use small-amplitude (a few to a dozen bases) 5′–3′ and 3′–5′ oscillations with a net 5′–3′ movement to search the AUG codon, and therefore, competition exists between closely spaced AUGs for translation initiation.
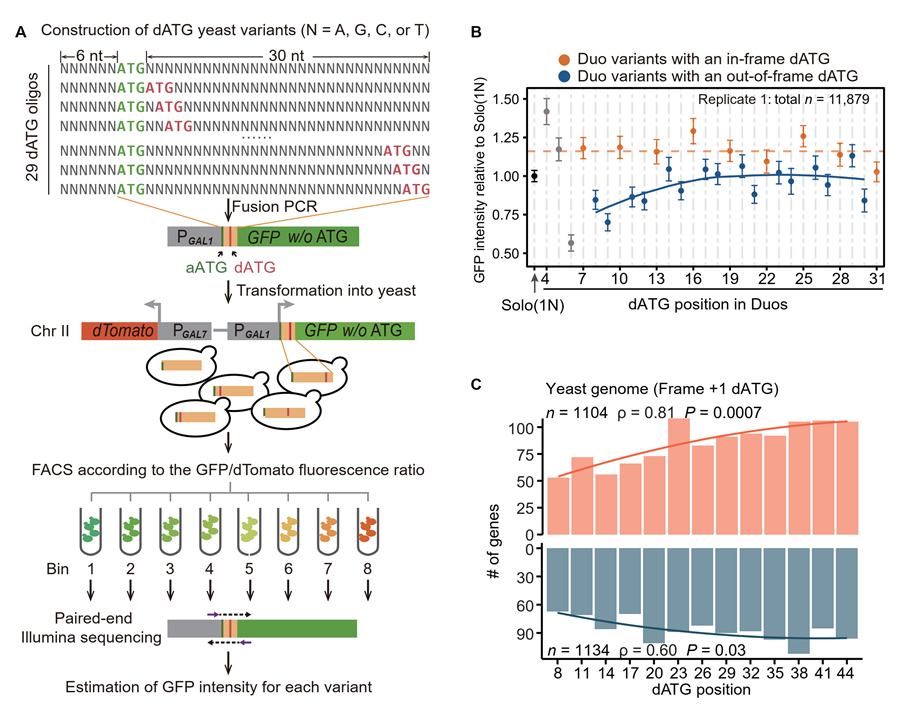
(A) High-throughput construction and phenotyping of a dual ATG library; (B) The inhibitory effects on translation initiation by proximal out-of-frame dATGs; (C) Distribution of out-of-frame dATG at different distances from aATG in yeast genome (top: “strong” context of dATGs, bottom: “weak” context of dATGs). (Image by IGDB)
The researchers initially observed that not only uAUGs but also the proximal out-of-frame downstream AUGs (dAUGs) can inhibit the translation initiation of the aAUG. In order to systematically investigate the role of dAUGs, the researchers constructed a dual ATG reporter library containing 13,437 yeast variants, each of which created a ATG triplet at a random position within the 30-nt downstream region of the aATG of a reporter gene, the green fluorescent protein (GFP). They then obtained the corresponding GFP level for each variant using a high-throughput approach. The results showed that out-of-frame dAUGs could inhibit translation initiation at the aAUG, but with diminishing strength over an increasing distance between aAUG and dAUG, undetectable beyond ~17 nt. These observations suggested that ribosomes frequently scan in the 3′–5′ direction since the ribosomes that could have initiated translation at aAUG appeared retained by a dAUG.
Consistent with the prediction of the bidirectional scanning model, the researchers also observed that the inhibitory effect of uAUG is position dependent: GFP intensities increased with decreasing uAUG-aAUG distance. The researchers further simulated the scanning process using a modified random walk model based on the massive GFP intensity data obtained in the high-throughput experiments, and estimated motion parameters of the scanning model using a Markov Chain Monte Carlo algorithm. The results showed that each triplet was on average scanned approximately ten times by the ribosome, resulting in a net leakage rate of 8% for an AUG triplet although the average leakage rate of every single scan for an AUG triplet was 77%.
The presence of proximal out-of-frame dATG may lead to reduced translation efficiency of functional proteins and increased synthesis of potentially cytotoxic peptides, an effect that should in turn affect the evolution of sequences downstream of aATG. The researchers then indeed observed that the number of out-of-frame dATGs increased gradually with distance from the aATG in yeast and human genomes, implying that the bidirectional scanning process is a general mechanism driving the evolution of eukaryotic genomes.
This study was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, and the State Key Laboratory of Plant Genomics.
Contact:
Professor QIAN Wenfeng
Institute of Genetics and Developmental Biology, Chinese Academy of Science
 (A) High-throughput construction and phenotyping of a dual ATG library; (B) The inhibitory effects on translation initiation by proximal out-of-frame dATGs; (C) Distribution of out-of-frame dATG at different distances from aATG in yeast genome (top: “strong” context of dATGs, bottom: “weak” context of dATGs). (Image by IGDB)The researchers initially observed that not only uAUGs but also the proximal out-of-frame downstream AUGs (dAUGs) can inhibit the translation initiation of the aAUG. In order to systematically investigate the role of dAUGs, the researchers constructed a dual ATG reporter library containing 13,437 yeast variants, each of which created a ATG triplet at a random position within the 30-nt downstream region of the aATG of a reporter gene, the green fluorescent protein (GFP). They then obtained the corresponding GFP level for each variant using a high-throughput approach. The results showed that out-of-frame dAUGs could inhibit translation initiation at the aAUG, but with diminishing strength over an increasing distance between aAUG and dAUG, undetectable beyond ~17 nt. These observations suggested that ribosomes frequently scan in the 3′–5′ direction since the ribosomes that could have initiated translation at aAUG appeared retained by a dAUG.Consistent with the prediction of the bidirectional scanning model, the researchers also observed that the inhibitory effect of uAUG is position dependent: GFP intensities increased with decreasing uAUG-aAUG distance. The researchers further simulated the scanning process using a modified random walk model based on the massive GFP intensity data obtained in the high-throughput experiments, and estimated motion parameters of the scanning model using a Markov Chain Monte Carlo algorithm. The results showed that each triplet was on average scanned approximately ten times by the ribosome, resulting in a net leakage rate of 8% for an AUG triplet although the average leakage rate of every single scan for an AUG triplet was 77%.The presence of proximal out-of-frame dATG may lead to reduced translation efficiency of functional proteins and increased synthesis of potentially cytotoxic peptides, an effect that should in turn affect the evolution of sequences downstream of aATG. The researchers then indeed observed that the number of out-of-frame dATGs increased gradually with distance from the aATG in yeast and human genomes, implying that the bidirectional scanning process is a general mechanism driving the evolution of eukaryotic genomes.This study was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, and the State Key Laboratory of Plant Genomics.Contact:Professor QIAN WenfengInstitute of Genetics and Developmental Biology, Chinese Academy of ScienceEmail: wfqian@genetics.ac.cn
(A) High-throughput construction and phenotyping of a dual ATG library; (B) The inhibitory effects on translation initiation by proximal out-of-frame dATGs; (C) Distribution of out-of-frame dATG at different distances from aATG in yeast genome (top: “strong” context of dATGs, bottom: “weak” context of dATGs). (Image by IGDB)The researchers initially observed that not only uAUGs but also the proximal out-of-frame downstream AUGs (dAUGs) can inhibit the translation initiation of the aAUG. In order to systematically investigate the role of dAUGs, the researchers constructed a dual ATG reporter library containing 13,437 yeast variants, each of which created a ATG triplet at a random position within the 30-nt downstream region of the aATG of a reporter gene, the green fluorescent protein (GFP). They then obtained the corresponding GFP level for each variant using a high-throughput approach. The results showed that out-of-frame dAUGs could inhibit translation initiation at the aAUG, but with diminishing strength over an increasing distance between aAUG and dAUG, undetectable beyond ~17 nt. These observations suggested that ribosomes frequently scan in the 3′–5′ direction since the ribosomes that could have initiated translation at aAUG appeared retained by a dAUG.Consistent with the prediction of the bidirectional scanning model, the researchers also observed that the inhibitory effect of uAUG is position dependent: GFP intensities increased with decreasing uAUG-aAUG distance. The researchers further simulated the scanning process using a modified random walk model based on the massive GFP intensity data obtained in the high-throughput experiments, and estimated motion parameters of the scanning model using a Markov Chain Monte Carlo algorithm. The results showed that each triplet was on average scanned approximately ten times by the ribosome, resulting in a net leakage rate of 8% for an AUG triplet although the average leakage rate of every single scan for an AUG triplet was 77%.The presence of proximal out-of-frame dATG may lead to reduced translation efficiency of functional proteins and increased synthesis of potentially cytotoxic peptides, an effect that should in turn affect the evolution of sequences downstream of aATG. The researchers then indeed observed that the number of out-of-frame dATGs increased gradually with distance from the aATG in yeast and human genomes, implying that the bidirectional scanning process is a general mechanism driving the evolution of eukaryotic genomes.This study was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, and the State Key Laboratory of Plant Genomics.Contact:Professor QIAN WenfengInstitute of Genetics and Developmental Biology, Chinese Academy of ScienceEmail: wfqian@genetics.ac.cn CAS
CAS
 中文
中文




.png)
