Polyploidy is prevalent in plants and has played a significant role in the evolution of almost all angiosperm genomes. For example, many domesticated crops are either polyploids (e.g. wheat, coffee, cotton and peanut) or paleopolyploids (e.g. rice, maize, soybean and sorghum). After polyploidization, particularly in allopolyploidy, the subgenomes may reduce potential genetic incompatibilities through rediploidization, a process that involves rapid gene losses. Despite the important role of diploidization in the evolutionary innovation of new genes and new species, the evolutionary forces that drive the diploidization process and their underlying mechanisms remain unclear.
Allotetraploid broomcorn millet (Panicum miliaceum L.), which belongs to the Paniceae tribe of the Gramineae family, is one of the earliest domesticated crops in North China. Broomcorn millet performs C4 photosynthesis, is highly adaptable and drought tolerant, and has a short growth period. It is an important source of food in some semi-arid areas and a valuable germplasm resource to help with the current global drought and food crisis. However, due to the ancestral diploid progenitors have not been determined, the subgenomes identification and the allotetraploidization time estimate are challenging in broomcorn millet, which limits the understanding of the origin and diploidization of this ancient allotetraploid crop.
In a recent study, Prof. CHEN Mingsheng’s group from the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS) de novo assembled a high-quality chromosome-scale genome sequence of the broomcorn millet.
They divided the broomcorn millet genome into two subgenomes using an evolutionary distance-based method using P. hallii as an intermediary. Successful identifying of the subgenomes provides a basis for studying allopolyploid evolution in broomcorn millet. Moreover, the approach to determine the subgenomes of an allopolyploid in this research is a reliable reference for studies of other allopolyploids without confirmed ancestral diploid progenitors.
Furthermore, they calculated the transposable elements (TEs) divergence between the broomcorn millet subgenomes to estimate the time of the allotetraploidization event. Their analyses revealed that the two subgenomes diverged at ~4.8 million years ago (Mya), while the allotetraploidization of broomcorn millet may have occurred about ~0.48 Mya, suggesting that broomcorn millet is a relatively recent allotetraploid.
Comparative analyses showed that subgenome B was larger than subgenome A in size, which was caused by the biased accumulation of long terminal repeat retrotransposons in the progenitor of subgenome B before polyploidization. Notably, the accumulation of biased mutations in the transposable element-rich subgenome B led to more gene losses.
Additionally, they revealed that none of the homoeologs co-expressed in both subgenomes and none of the homoeologs expressed in one subgenome showed evidence of subgenome expression dominance. However, they found the minimally expressed genes in P. hallii tended to be lost during diploidization of broomcorn millet.
This study presents an example of diploidization, in which the tetraploid genome underwent subtle biased fractionation; however, no subgenome expression dominance was established.
Results entitled “Biased mutations and gene losses underlying diploidization of the tetraploid broomcorn millet genome” was published in The Plant Journal and it was supported by National Natural Science Foundation of China and The State Key Laboratory of Plant Genomics.
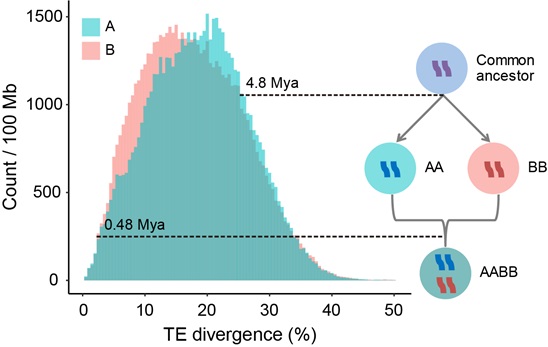 Distribution of divergence of the transposons and a model of tetraploid formation in broomcorn millet. The y-axis represents the TE counts normalized by subgenome size. Model of tetraploid formation and evolution of transposon in the subgenomes: AA and BB diverged from a common ancestor. Due to their different environments, the evolutionary rates of the transposons in the genomes were also different, which led to differences in the distribution of their transposons divergences. Then, AA and BB merged into a tetraploid, and the A and B subgenomes in the newly formed tetraploid genome were in the same environment, so the evolutionary rates of transposons tended to be the same. (Image by IGDB)
Distribution of divergence of the transposons and a model of tetraploid formation in broomcorn millet. The y-axis represents the TE counts normalized by subgenome size. Model of tetraploid formation and evolution of transposon in the subgenomes: AA and BB diverged from a common ancestor. Due to their different environments, the evolutionary rates of the transposons in the genomes were also different, which led to differences in the distribution of their transposons divergences. Then, AA and BB merged into a tetraploid, and the A and B subgenomes in the newly formed tetraploid genome were in the same environment, so the evolutionary rates of transposons tended to be the same. (Image by IGDB)
Contact:
Prof. CHEN Mingsheng
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences

 CAS
CAS
 中文
中文




.png)
