GAO Caixia’s group from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences (CAS) has developed a new method of downregulating gene translation to a predictable and desired level in plants by precisely engineering upstream open reading frames (uORFs).
The study was published online in Nature Biotechnology on Mar. 9.
The development and application of genome editing in plants has revolutionized molecular design-based crop breeding. Developing methods for fine-tuning gene expression based on precise genome changes is crucial for breeding new and desired traits into crops. Widely used gene editing tools such as CRISPR-Cas, CRISPR interference and RNA interference generally completely prevent gene transcription or reduce it to a defined but unpredictable level.
CRISPR-Cas9-driven mutagenesis of promoters provides a method for generating quantitative phenotypic changes at the transcriptional level and producing a wide range of gene expression levels. A method for predictably and incrementally regulating endogenous gene expression at the translational level would further expand on methods of controlling gene activity.
In 2018, GAO’s group led the development of an efficient and tunable method for upregulating protein expression by knocking out endogenous uORFs. In 2020, they applied this technology to the genetic improvement of strawberries, generating a variety of new strawberry germplasms with different sugar content.
uORFs are important and universal cis-regulatory elements in eukaryotes that are thought to be associated with reduced mRNA translation. Many factors, such as uORF length and the distance between uORFs and primary open reading frames (pORFs), affect the inhibitory activity of uORFs. Therefore, the researchers proposed two strategies for downregulating gene translation: introducing de novo uORFs at the 5’ untranslated region (5’ UTR) of a target gene and mutating stop codons of endogenous uORFs to extend their coding sequences and enhance their inhibitory ability.
Dual-luciferase and western blot assays in transient systems showed that the newly produced and extended uORFs downregulated LUC/REN activity ratios to 9.5-86.9% of wildtype levels but had no effect on LUC/REN mRNA levels, thereby demonstrating translational control.
Using precise genome editing technologies such as base editors and prime editors, the researchers obtained mutant rice plants carrying new and extended uORFs. They then confirmed that the effects of these engineered uORFs on phenotypes and protein expression levels were the same as those observed in the transient reporter system.
To incrementally downregulate the translation of genes, the researchers combined the above approaches and generated a series of uORFs with different inhibitory capacities at the 5’ UTRs of OsTCP19, OsTB1 and OsDLT. Transient systems showed that these uORFs incrementally downregulated the translation of pORFs to 2.5-84.9% of the wildtype level.
In addition, by editing the 5’ UTR of rice OsDLT, which encodes for DWARF AND LOW-TILLERING—a member of the GRAS family of transcriptional regulators involved in the brassinosteroid (BR) transduction pathway—they obtained a range of plants with different BR sensitivity, plant height and number of tillers that matched the corresponding reduction in OsDLT levels observed in a transient protoplast assay.
By engineering uORFs, the researchers have developed an efficient and widely applicable method for downregulating gene translation to predictable and desired levels in plants, which will greatly improve future crop breeding.
This study was supported by grants from the National Key Research and Development Program, the Strategic Priority Research Program of the CAS, and the Ministry of Agriculture and Rural Affairs of China.
Figure: Breeding plants with a predicted phenotype by engineering uORFs (Image by IGDB). (a) Repressed protein expression by generating de novo uORFs. (b) Repressed protein expression by extending endogenous uORFs. (c) Downregulating protein expression in a graded fashion by producing uORFs with diverse inhibitory activities. (d, e) BR sensitivity (d) and morphology (e) of wild type (WT) plants and T1 progenies of edited plants carrying the engineered uORFs.
Contact:
Dr. GAO Caixia
Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences
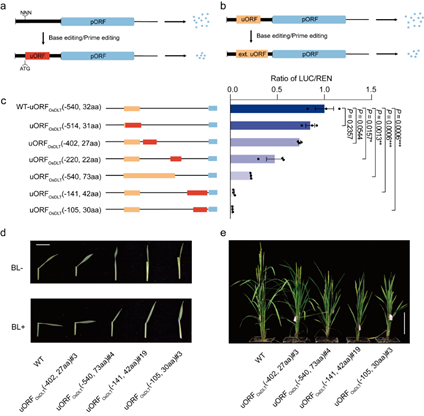 Figure: Breeding plants with a predicted phenotype by engineering uORFs (Image by IGDB). (a) Repressed protein expression by generating de novo uORFs. (b) Repressed protein expression by extending endogenous uORFs. (c) Downregulating protein expression in a graded fashion by producing uORFs with diverse inhibitory activities. (d, e) BR sensitivity (d) and morphology (e) of wild type (WT) plants and T1 progenies of edited plants carrying the engineered uORFs.Contact:Dr. GAO CaixiaInstitute of Genetics and Developmental Biology of the Chinese Academy of SciencesEmail: cxgao@genetics.ac.cn
Figure: Breeding plants with a predicted phenotype by engineering uORFs (Image by IGDB). (a) Repressed protein expression by generating de novo uORFs. (b) Repressed protein expression by extending endogenous uORFs. (c) Downregulating protein expression in a graded fashion by producing uORFs with diverse inhibitory activities. (d, e) BR sensitivity (d) and morphology (e) of wild type (WT) plants and T1 progenies of edited plants carrying the engineered uORFs.Contact:Dr. GAO CaixiaInstitute of Genetics and Developmental Biology of the Chinese Academy of SciencesEmail: cxgao@genetics.ac.cn CAS
CAS
 中文
中文




.png)
