Center for Crop Phenomics from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences and Prof. YANG Wanneng group from Huazhong Agricultural University developed a strategy for acquiring and analyzing 58 image-based traits (i-traits) during the whole growth period of rice. The study was published in Plant Phenomics (DOI: 10.34133/plantphenomics.0058).
In this study, on the basis of the i-trait phenomes obtained, up to 84.8% of the phenotypic variance in the rice yield could be explained by 8 traits among all i-traits considered, and these traits could thus be applied to predict the final rice yield better than predictions performed with other trait groups. This finding indicates that the i-traits obtained here can comprehensively reflect the growth status of rice, with the exception of the final yield.
To further explore the good environmental adaptability of the crop growth and development model, which would also show high inosculation with the latitude of the breeding region, the breeding region was divided into different latitude levels. Interestingly, in addition to the HS of japonica rice showing an increasing negative correlation with the region latitude, some traits showed correlations with the regional latitude with significant differences among rice accession groups, such as PanicleTPA and TFN of indica rice, which may explain the difference in the yields of different rice cultivars with the difference in the geographical environment of the breeding region. Moreover, the differences in rice subspecies were analyzed using the image-based phenotypic traits, which, in addition to PlantYpar, indicated the senescence degree at the whole-plant and leaf level, and the HS, AveLW, and PanicleYPA terms also showed significant differences.
Furthermore, PCA was also performed for the characteristics of the rice accessions in the organ–temporal dimension, and GWAS using PC scores was utilized to identify genetic factors. Among the PC values of the organ and temporal traits, a QTL on chromosome 3, with a lead SNP on chromosome 3: 24262598, was detected by both the organ and temporal PCs, indicating that this locus is deserving of further studies in the future involving confirmations with larger panels. As a proof of concept, this study demonstrates the potential value of this pathway in high-throughput phenomics studies and provides a reference for further similar studies in the future.
This work was supported by the National Key Research and Development Plan, Strategic Priority Research Program of the Chinese Academy of Sciences, National Natural Science Foundation of China Key Projects of Natural Science Foundation of Hubei Province, Major Science and Technology Project of Hubei Province, and Fundamental Research Funds for the Central Universities.
Figure: High-throughput rice plant phenotyping results (Image by IGDB). (A) Images of rice acquisitions. (B) Dynamic shoot growth during all growth stages. (C) Group clustering and regional division of different rice genotypes. (D) Data analysis of i-traits.
Contact:
Dr. HU Weijuan
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: wjhu@genetics.ac.cn
Dr. YANG Wanneng
National Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Hubei Hongshan Laboratory, Huazhong Agricultural University
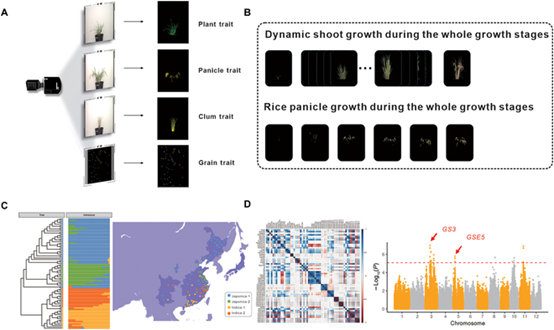 Figure: High-throughput rice plant phenotyping results (Image by IGDB). (A) Images of rice acquisitions. (B) Dynamic shoot growth during all growth stages. (C) Group clustering and regional division of different rice genotypes. (D) Data analysis of i-traits.Contact:Dr. HU WeijuanInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: wjhu@genetics.ac.cnDr. YANG WannengNational Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Hubei Hongshan Laboratory, Huazhong Agricultural UniversityEmail: ywn@mail.hzau.edu.cn
Figure: High-throughput rice plant phenotyping results (Image by IGDB). (A) Images of rice acquisitions. (B) Dynamic shoot growth during all growth stages. (C) Group clustering and regional division of different rice genotypes. (D) Data analysis of i-traits.Contact:Dr. HU WeijuanInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: wjhu@genetics.ac.cnDr. YANG WannengNational Key Laboratory of Crop Genetic Improvement, National Center of Plant Gene Research (Wuhan), Hubei Hongshan Laboratory, Huazhong Agricultural UniversityEmail: ywn@mail.hzau.edu.cn CAS
CAS
 中文
中文




.png)
