Soil salinization reduces soil fertility and crop yields, posing a major threat to global agriculture. The Salt Overly Sensitive (SOS) signaling pathway, consisting of key components SOS1(a plasma membrane-localized Na+/H+ antiporter), SOS2 (a serine/threonine protein kinase) and SOS3(a calcium-binding protein), plays a central role in conferring salt tolerance in plants. SOS1 is the sole known plasma membrane Na+ efflux protein in plants to date, and its crucial role in maintaining ion homeostasis has been firmly established through genetic, biochemical, and physiological research conducted in both monocots and dicots. However, the molecular mechanism governing the function of SOS1 has remained largely enigmatic.
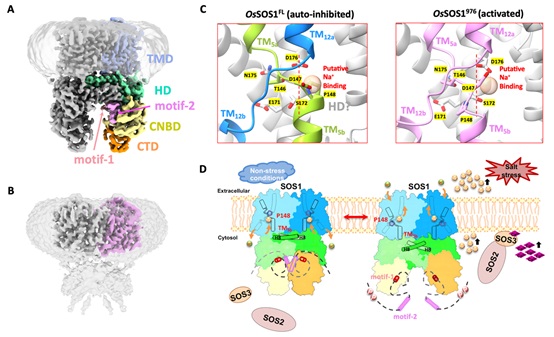
Recently, Dr. CHEN Yu-hang’s team solved the cryo-EM structures of rice SOS1 (OsSOS1) from Oryza sativa, both in its full-length auto-inhibited state (OsSOS1FL, Figure A) and its activated state (OsSOS1976, a constitutively active truncated version lacking the C-terminal auto-inhibitory domain, Figure B). The OsSOS1FL structure revealed that SOS1 forms a homodimer, with each protomer featuring an NhaA-fold transmembrane domain (TMD) and three cytosolic regulatory domains: a helical domain (HD), a cyclic nucleotide-binding domain (CNBD), and a C-terminal β-roll domain (CTD). Two highly conserved auto-inhibitory motifs (motif-1 and motif-2) bind to the cytosolic regulatory domains, forming an auto-inhibited conformation. In contrast, the OsSOS1976 structure showed that its scaffold domain and transport domain undergo an elevator-like "up/down" movement, accompanied by the downward displacement of the TM5b segment and the re-positioning of the gating residue Pro148.
Based on in-depth structural analysis, researchers have proposed the working model of SOS1. In the absence of salt stress, the cytosolic regulatory domains of SOS1 are stabilized by binding to the conserved auto-inhibitory motif in its pre-activation state. The helical pair (helices H8 and H9) within the HD domain closely interacts with TM5b, thus positioning Pro148 at its occluded position to prevent Na+ binding and maintaining an auto-inhibited state. When plants encounter salt stress, the elevated calcium signal is sensed by SOS3, which forms a complex with SOS2 to activate its kinase activity. This, in turn, phosphorylates the Na+/H+ antiporter SOS1. Phosphorylation of SOS1 leads to the release of its auto-inhibition, causing drastic conformational changes in the cytosolic regulatory domains. Particularly, it disengages the TM5b segment from its interaction with the helical pair (H8/H9) of the HD domain, resulting in its downward movement and unwinding of motif 144SATDP148, accompanied by re-positioning Pro148 from Na+ occluded position to Na+ transporting position, Figure C). These findings explain how SOS1 switches from a resting autoinhibitory state to an activated state, providing a structural basis for understanding the molecular mechanism of SOS1 activation (Figure D).
The research findings, entitled "Structure and activation mechanism of the rice Salt Overly Sensitive 1 (SOS1) Na
+/H
+ antiporter " (DOI: 10.1038/s41477-023-01551-5), together with a research briefing "Structural insight into SOS signaling in response to salt stress" (DOI:
https://doi.org/10.1038/s41477-023-01553-3) were published online in the
Nature Plants on October 26, 2023. This research was funded by the Strategic Priority Research Program of CAS and the National Key R&D Program of China.
Cryo-EM structures and activation mechanism of rice Na+/H+ antiporter SOS1 (Image by IGDB)
A. Cryo-EM structure of full-length rice SOS1 in the auto-inhibited state (OsSOS1FL, residue 1-1146); B. Cryo-EM structure of the truncated SOS1 in the constitutively activated state (OsSOS1976, residue 1-976); C. Comparison of the gating Pro148 in the Na+/H+ ion-transport pathway: Na+-occluded position (In OsSOS1FL, left) and Na+-transporting position (In OsSOS1976, right); D. Molecular mechanism of SOS1 activation in response to salt stress.
Contact:
Dr. CHEN Yu-hang
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
 Cryo-EM structures and activation mechanism of rice Na+/H+ antiporter SOS1 (Image by IGDB)A. Cryo-EM structure of full-length rice SOS1 in the auto-inhibited state (OsSOS1FL, residue 1-1146); B. Cryo-EM structure of the truncated SOS1 in the constitutively activated state (OsSOS1976, residue 1-976); C. Comparison of the gating Pro148 in the Na+/H+ ion-transport pathway: Na+-occluded position (In OsSOS1FL, left) and Na+-transporting position (In OsSOS1976, right); D. Molecular mechanism of SOS1 activation in response to salt stress.Contact:Dr. CHEN Yu-hangInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesE-mail: yuhang.chen@genetics.ac.cn
Cryo-EM structures and activation mechanism of rice Na+/H+ antiporter SOS1 (Image by IGDB)A. Cryo-EM structure of full-length rice SOS1 in the auto-inhibited state (OsSOS1FL, residue 1-1146); B. Cryo-EM structure of the truncated SOS1 in the constitutively activated state (OsSOS1976, residue 1-976); C. Comparison of the gating Pro148 in the Na+/H+ ion-transport pathway: Na+-occluded position (In OsSOS1FL, left) and Na+-transporting position (In OsSOS1976, right); D. Molecular mechanism of SOS1 activation in response to salt stress.Contact:Dr. CHEN Yu-hangInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesE-mail: yuhang.chen@genetics.ac.cn CAS
CAS
 中文
中文




.png)
