Structural variations (SVs) are widespread in plant genomes and play a crucial role in gene expression regulation, phenotype formation, and adaptive evolution. Due to their large span and structural complexity, accurately detecting SVs is highly challenging. In recent years, advancements in third-generation sequencing have significantly improved sequencing length and accuracy, providing an opportunity for precise genome-wide SV detection. However, most SV analysis algorithms and software are designed and developed for the human genome, and their applicability to complex plant genomes has yet to be evaluated. Therefore, conducting benchmark tests on SV detection algorithms for plant genomes is of great importance in uncovering the mechanisms of SVs.
On September 6, 2024, the research group led by LU Fei at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, published a paper titled "Structural variation discovery in wheat using PacBio high-fidelity sequencing" in The Plant Journal. This study focused on allohexaploid bread wheat and its ancestral donors, using PacBio high-fidelity (HiFi) sequencing data to conduct a benchmark test of third-generation sequencing alignment algorithms and structural variation detection algorithms (Figure 1).
The results showed that for deletions, the main factor affecting detection accuracy (F-score) was the structural variation detection software, explaining 87.73% of the total variance in accuracy. For insertions, both the third-generation sequencing alignment software and the structural variation detection software significantly contributed to detection accuracy, accounting for 38.25% and 49.32% of the total variance, respectively. Among the third-generation alignment software, Winnowmap2 and NGMLR were best suited for detecting deletions and insertions, respectively, while the structural variation detection software SVIM performed best in detecting both types of variants.
This combination of alignment and detection software represents the most effective method for detecting structural variations in wheat. Additionally, the study confirmed that low-coverage PacBio HiFi (0.3X) third-generation sequencing data can also accurately detect genome structural variations.
This study provides the most optimal analytical workflow for detecting structural variations in the wheat genome and demonstrates the capability of low-coverage PacBio HiFi third-generation sequencing in detecting structural variations, offering theoretical and technical support for large-scale population studies of structural variations.
The study was supported by the National Key R&D Program, the National Natural Science Foundation of China, the Chinese Academy of Sciences Strategic Priority Research Program, the "Revealing the Champion" project of the Yazhou Bay Seed Lab in Hainan, and the Open Project of the State Key Laboratory of Plant Cell and Chromosome Engineering.
Figure 1: Overview of benchmark testing for genome structural variation detection algorithms (Image by IGDB)
Contact:
Dr. LU Fei
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: flu@genetics.ac.cn
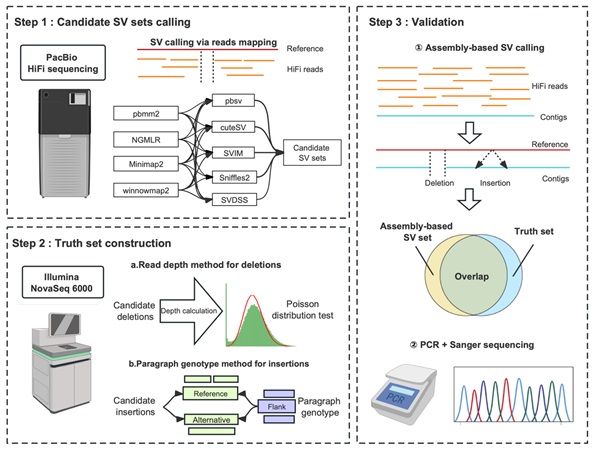 Figure 1: Overview of benchmark testing for genome structural variation detection algorithms (Image by IGDB)Contact:Dr. LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: flu@genetics.ac.cn
Figure 1: Overview of benchmark testing for genome structural variation detection algorithms (Image by IGDB)Contact:Dr. LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: flu@genetics.ac.cn CAS
CAS
 中文
中文




.png)
