To tackle these challenges, a team led by Professor WANG Yanpeng from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences, in collaboration with the Wheat Genetics and Genomics Center of China Agricultural University, develop a precise and efficient scarless large DNA fragments technology in plants, which can efficiently facilitate precise deletion, replacement, and inversion of large DNA fragments in plants. This study was published in
Nature Plants (
https://doi.org/10.1038/s41477-024-01898-3).
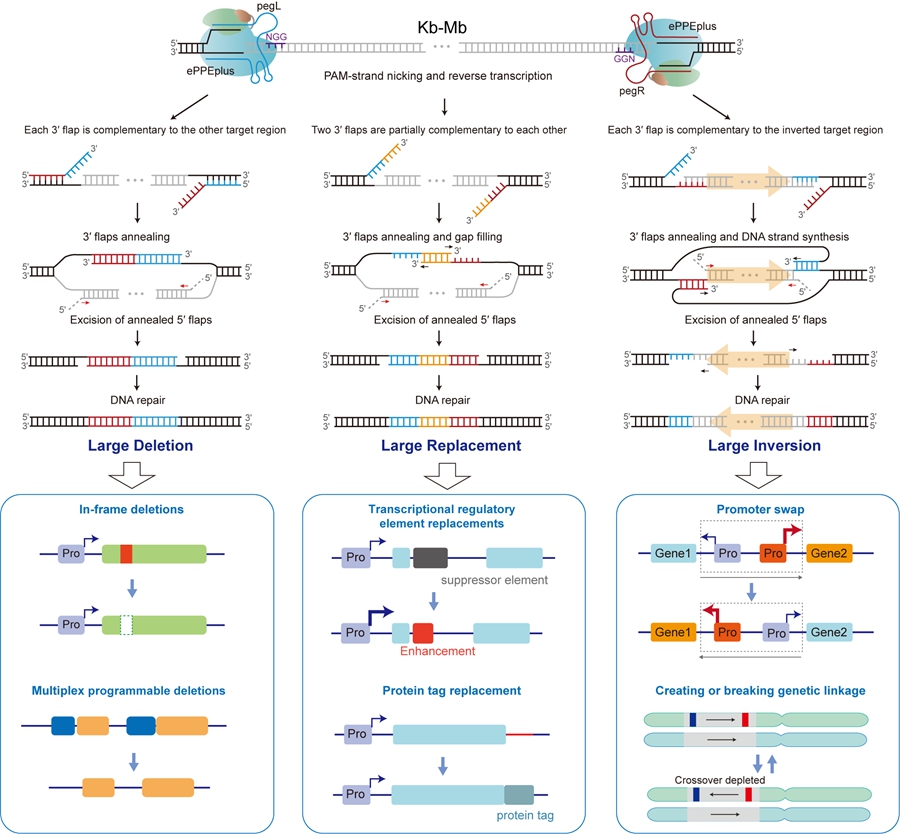
This study used an optimized prime editor, ePPEplus, which contains a V223A mutation in the reverse transcriptase and the R221K and N394K mutations in nCas9 (H840A), with the dual epegRNAs to target complementary DNA strands with a PAM-in orientation. The ePPEplus-epegRNA complex recognizes the target sites adjacent to the large DNA fragment being edited and nick the PAM-containing DNA strands, respectively. The 3′ end of the nicking sites hybridize to the corresponding PBS of epegRNA and then reverse transcribe RT templates (RTTs), generating two 3′ DNA flaps at the respective target sites. Theoretically, as depicted in Figure, through the manipulating of these3′ DNA flaps, it is possible to achieve large DNA deletions, replacements, or inversions.
Here are the possible scenarios: (1) If each 3′ flap sequence is complementary to the region targeted by the other pegRNA, the two 3′ flaps will anneal to form double-stranded DNA, resulting in the excision of the original DNA strands (5′ flaps). The subsequent DNA repair process leads to the precise deletion of DNA fragments between the two pegRNAs. (2) In cases where the two 3′ flaps generated by the RTT are partially complementary to each other and also contain a desired insertion sequence, two 3′ flaps annealing, followed by DNA synthesis and excision of the 5′ flaps, results in a deletion and simultaneous insertion of the desired sequence between the two pegRNAs. (3)
When each 3′ flap sequence is complementary to the inverted region targeted by the other pegRNA, each 3′ flap will anneal to its corresponding sequence. This annealing will serve as a primer to initiate DNA synthesis in the opposite orientation.
Subsequently, the 5′ flaps will be excised, followed by ligation and DNA repair, which may result in an inversion. These three scenarios represent the genome-editing approach that we term dual prime editing (DualPE).
DualPE thus provides a precise and efficient approach for large DNA sequence and chromosomal engineering, expanding the availability of precision genome editing for crop improvement.
DualPE-mediated large DNA deletions, replacements, and inversions in plant (Image by IGDB)
Contact:
Prof. WANG Yanpeng
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: yanpengwang@genetics.ac.cn
 DualPE-mediated large DNA deletions, replacements, and inversions in plant (Image by IGDB)Contact:Prof. WANG YanpengInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: yanpengwang@genetics.ac.cn
DualPE-mediated large DNA deletions, replacements, and inversions in plant (Image by IGDB)Contact:Prof. WANG YanpengInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: yanpengwang@genetics.ac.cn CAS
CAS
 中文
中文




.png)
