Wheat (Triticum aestivum L.) is a key global food crop, but its complex, highly repetitive allohexaploid genome has made complete sequencing a challenge. In 2018, the International Wheat Genome Sequencing Consortium (IWGSC) released the Chinese Spring (CS) wheat reference genome, which has since been pivotal for wheat research. However, many repetitive regions and complex structures remain unannotated, hindering in-depth genomic analysis.
On February 13, 2025, a team led by FU Xiangdong and LU Fei from the Chinese Academy of Sciences, in collaboration with China Agricultural University, published a paper in
Molecular Plant (
https://doi.org/10.1016/j.molp.2025.02.002) titled "Near-complete assembly and comprehensive annotation of the wheat Chinese Spring genome."
This study achieved a near-complete assembly of the wheat CS genome (CS-CAU) using Oxford Nanopore Technologies (ONT) ultra-long read sequencing, PacBio HiFi high-accuracy sequencing, and Hi-C data. The CS-CAU genome spans 14.46 Gb with over 99.996% base accuracy, and just 290 gaps remain, primarily due to ultra-long tandem repeats. Notably, chromosomes 1D, 3D, 4D, and 5D were assembled without gaps for the first time, with chromosomes 1D and 5D reaching the telomere-to-telomere (T2T) level. This breakthrough addresses high-repetition sequences and polyploidy issues in wheat genomics, offering a model for sequencing other complex crop genomes.
The study also annotated 151,405 high-confidence genes, including 59,180 newly identified genes, 7,602 of which were assembled for the first time. This is crucial for wheat gene function research.
The research team also resolved the distribution and expression of six classes of seed storage proteins (SSPs) across the genome. The ω-gliadin expression is entirely from the B sub-genome, while other SSPs, including α/γ-gliadin, ALP, and glutenins, are predominantly expressed from the D sub-genome. These findings are vital for understanding and improving wheat gluten quality.
Additionally, centromeric regions were mostly assembled, with the exception of the ultra-long GAA repeat sequence in chr1B. These centromeres are rich in retrotransposons, particularly CRW and Quinta, which vary in distribution across the A/B and D sub-genomes. The study also explored the historical expansion of these retrotransposons.
This study utilized Oxford Nanopore, PacBio HiFi sequencing, and Hi-C technology to create a near-complete assembly of the Chinese Spring wheat genome (CS-CAU), advancing wheat genetic research.
This work was supported by the National Key Research and Development Program of China, the National Natural Science Foundation of China, the Pinduoduo-China Agricultural University Research Fund, the New Cornerstone Investigation Program, and the 2115 Talent Development Program of China Agricultural University.
Figure 1. Near-complete assembly of the Chinese Spring genome. (Image by IGDB)
Contact:
Drs. FU Xiangdong and LU Fei
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
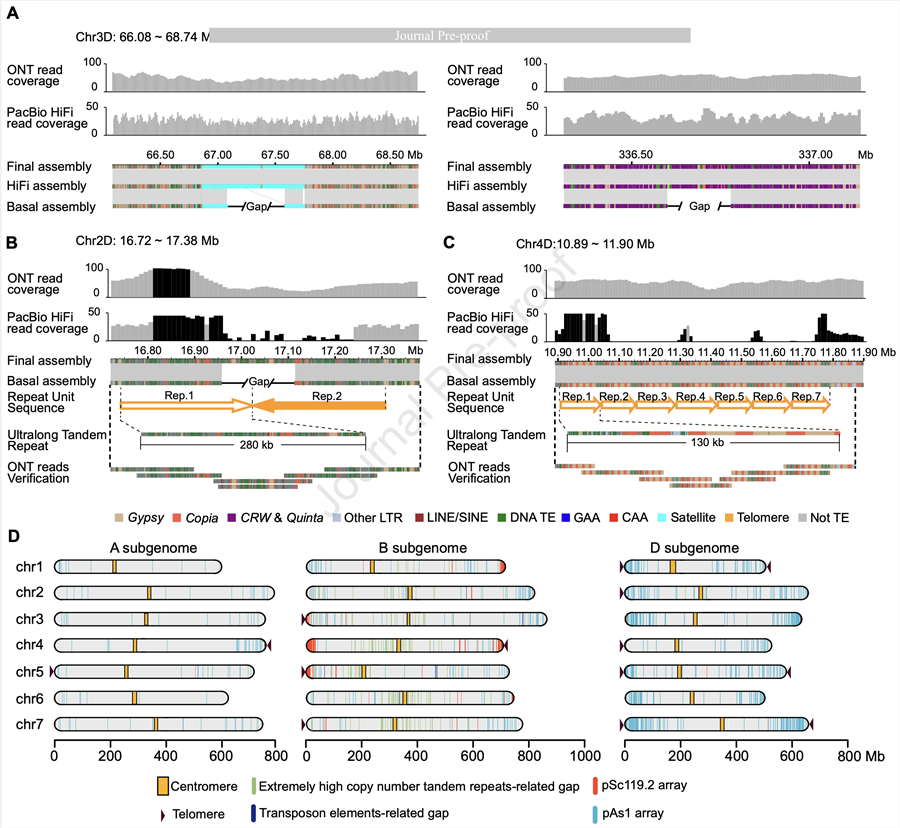 Figure 1. Near-complete assembly of the Chinese Spring genome. (Image by IGDB)Contact:Drs. FU Xiangdong and LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmails: xdfu@genetics.ac.cn, flu@genetics.ac.cn
Figure 1. Near-complete assembly of the Chinese Spring genome. (Image by IGDB)Contact:Drs. FU Xiangdong and LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmails: xdfu@genetics.ac.cn, flu@genetics.ac.cn CAS
CAS
 中文
中文




.png)
