Soybean is an important crop and a key source of plant-based protein and oil for humans and livestock. With global demand steadily increasing, it is projected that soybean production will need to double by 2050 compared to 2015 levels. Therefore, accelerating soybean breeding through molecular design strategies has become crucial. Among these efforts, uncovering key genes and their regulatory networks in soybean organ development is particularly vital for precise molecular breeding.
The research team led by TIAN Zhixi from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, in collaboration with partners, has pioneered a groundbreaking "macro-single-cell-spatial" three-level transcriptional analysis system.
They utilized Bulk RNA-seq data from 314 full-organ samples to precisely identify the developmental stages of soybean organs and their characteristic genes. Then they employed single-cell RNA sequencing (snRNA-seq) to capture cell-level expression profiles of five major functional organs (roots, nodules, shoot apices, leaves, and stems). Finally, using Stereo-seq technology, they visualized the three-dimensional spatial location information of gene expression, achieving for the first time a 3D visualization of gene expression in soybean organs.
By integrating data from various types of transcriptomic analyses, their study has comprehensively revealed the spatiotemporal dynamics of soybean gene expression, providing unprecedented insights into soybean development.
They identified organ-specific genes in soybeans, with root nodule-specific genes as a case study. It was confirmed that GmPMTs regulate nodule development through gene tandem duplication and expansion.
They also constructed an organ developmental atlas, focusing on leaf development. For the first time, transcriptional specificity was observed during the leaf expansion stage, and key long-chain fatty acid co-expression modules were identified, offering new clues for soybean leaf improvement.
Additionally, the development of a 3D spatial transcriptomic map of root tips, reveals the dynamic pathways of cell differentiation in soybean roots and provides a developmental blueprint for legume root research.
The researchers analyzed the cellular heterogeneity in root nodules using 3D single-cell spatial mapping, uncovering the spatial localization of symbiotic genes in root nodules. This establishes a cellular-level spatiotemporal coordinate system for studying root-microbe interactions.
Figure 1: The transcriptome map of soybean organ development. (Image by IGDB)
To democratize access to these insights, the team launched two public databases:
SoyOmics Transcriptome Atlas (https://ngdc.cncb.ac.cn/soyomics/index) and
SOTA: Soybean Organ Transcriptomic Atlas (
https://db.cngb.org/stomics/soybean). These platforms enable researchers worldwide to retrieve the expression characteristics of target genes during different developmental stages, in specific organs, and across various cell types, and accelerate functional gene research.
The "spatiotemporal atlas" constructed in this study comprehensively reveals the dynamic trajectories of gene expression during soybean growth and development, providing core technical support for understanding the mechanisms of soybean organ formation and mining key developmental regulatory genes.
The work titled “A large-scale integrated transcriptomic atlas for soybean organ development” was published in Molecular Plant (https://doi.org/10.1016/j.molp.2025.02.003.).
This work was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Taishan Scholars Program and the Xplorer Prize.
Contact:
TIAN Zhixi
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Yazhouwan National Laboratory
Email: tianzhixi@yzwlab.cn
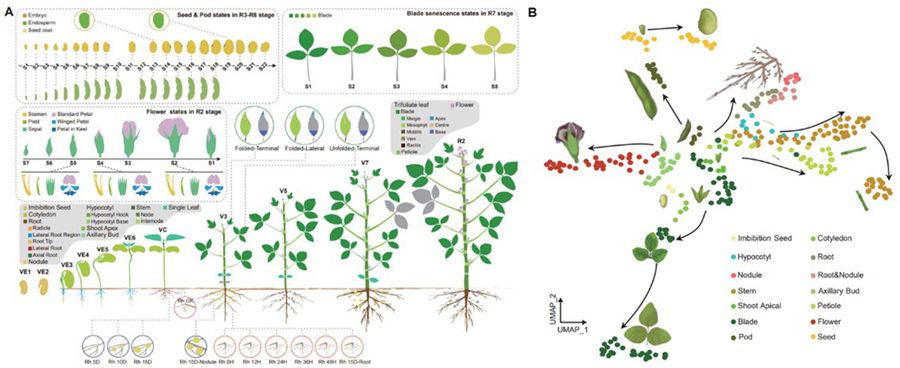 Figure 1: The transcriptome map of soybean organ development. (Image by IGDB)To democratize access to these insights, the team launched two public databases: SoyOmics Transcriptome Atlas (https://ngdc.cncb.ac.cn/soyomics/index) and SOTA: Soybean Organ Transcriptomic Atlas (https://db.cngb.org/stomics/soybean). These platforms enable researchers worldwide to retrieve the expression characteristics of target genes during different developmental stages, in specific organs, and across various cell types, and accelerate functional gene research.The "spatiotemporal atlas" constructed in this study comprehensively reveals the dynamic trajectories of gene expression during soybean growth and development, providing core technical support for understanding the mechanisms of soybean organ formation and mining key developmental regulatory genes.The work titled “A large-scale integrated transcriptomic atlas for soybean organ development” was published in Molecular Plant (https://doi.org/10.1016/j.molp.2025.02.003.).This work was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Taishan Scholars Program and the Xplorer Prize.Contact:TIAN ZhixiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesYazhouwan National LaboratoryEmail: tianzhixi@yzwlab.cn
Figure 1: The transcriptome map of soybean organ development. (Image by IGDB)To democratize access to these insights, the team launched two public databases: SoyOmics Transcriptome Atlas (https://ngdc.cncb.ac.cn/soyomics/index) and SOTA: Soybean Organ Transcriptomic Atlas (https://db.cngb.org/stomics/soybean). These platforms enable researchers worldwide to retrieve the expression characteristics of target genes during different developmental stages, in specific organs, and across various cell types, and accelerate functional gene research.The "spatiotemporal atlas" constructed in this study comprehensively reveals the dynamic trajectories of gene expression during soybean growth and development, providing core technical support for understanding the mechanisms of soybean organ formation and mining key developmental regulatory genes.The work titled “A large-scale integrated transcriptomic atlas for soybean organ development” was published in Molecular Plant (https://doi.org/10.1016/j.molp.2025.02.003.).This work was supported by the National Natural Science Foundation of China, the National Key Research and Development Program of China, the Taishan Scholars Program and the Xplorer Prize.Contact:TIAN ZhixiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesYazhouwan National LaboratoryEmail: tianzhixi@yzwlab.cn CAS
CAS
 中文
中文




.png)
