A collaborative research led by Dr. FENG Jian and Dr. HAN Fangpu at the Institute of Genetics and Developmental Biology (IGDB), Chinese Academy of Sciences, has achieved a major milestone in legume genomics by generating gap-free telomere-to-telomere (T2T) genome assemblies for two model Medicago species. This study provides unprecedented insights into the structure and evolution of plant centromeres, offering a valuable reference for legume functional genomics and precision breeding.
The findings, published on 25 July in the journal Molecular Plant (DOI:10.1016/j.molp.2025.07.016), are detailed in the article titled “Two complete telomere-to-telomere Medicago genomes reveal the landscape and evolution of centromeres.”
A breakthrough in legume genomics
Medicago is a key model genus in legume biology and includes major forage crops such as alfalfa. Its roots form nitrogen-fixing nodules through symbiosis with rhizobia, enhancing both plant growth and soil fertility. To fully understand the genetic and structural basis of these traits, complete and accurate genome assemblies are essential.
Using a combination of PacBio HiFi sequencing, ultra-long Oxford Nanopore reads, and Hi-C chromatin conformation capture, the team successfully constructed T2T genome assemblies for Medicago truncatula Jemalong A17 (A17 v6.0) and Medicago littoralis R108 (R108 v3.0). These assemblies span 494.47 Mb and 415.27 Mb, respectively, with >99% completeness based on BUSCO assessment, representing the most complete genomes available for the Medicago genus to date.
Figure. Phenotypicvariations observed between A17 and R108 (Image by IGDB)
Decoding the centromere: diversity and evolution
The researchers turned their attention to centromere, one of the most enigmatic regions of eukaryotic genomes, which plays a central role in chromosome segregation. Despite its functional importance, centromeric DNA has been historically difficult to assemble due to its highly repetitive nature. By integrating chromatin immunoprecipitation sequencing of the centromere-specific histone CENH3, fluorescence in situ hybridization (FISH), and de novo repeat annotation, the team discovered striking differences between the centromeres of A17 and R108. In A17, centromeres are primarily composed of two tandem satellite repeats: CentM168 and the species-specific CentM183. In contrast, R108 centromeres consist almost exclusively of CentM168, highlighting a distinct evolutionary trajectory between M. truncatula and M. littoralis.
This divergence illustrates rapid centromere evolution, even between closely related species. The researchers propose a model in which active centromeres are maintained by CENH3-enriched CentM168 arrays, while non-active pericentromeric domains are buffered by satellite-rich flanking regions, possibly reducing chromosomal instability.
In addition, they identified several highly chromosome-specific satellite repeats (e.g., CentM51, CentM515, CentM287), further underscoring the dynamic and mosaic nature of centromere evolution. Intriguingly, centromeric regions were also enriched in young long terminal repeat (LTR) retrotransposons, suggesting a potential role for transposable elements in shaping centromeric architecture and function.
Implications and future directions
The corresponding author, Dr. FENG Jian, said, “This study represents a major leap forward in our understanding of genome architecture and centromere evolution in legumes. It not only provides a high-quality genomic foundation for future gene discovery and trait mapping, but also sets the stage for engineering plant chromosomes with improved stability and transmission fidelity.”
Dr. HAN Fangpu, the co-corresponding author and an expert in plant cytogenetics, added, “For decades, the centromere has been a genomic blind spot. Now, with complete assemblies and detailed repeat maps, we can begin to ask new questions about how centromeres originate, diversify, and impact legume genome evolution.”
The T2T Medicago genomes generated in this work are publicly available and expected to accelerate research across legume biology, from symbiotic signaling and stress adaptation to chromosome engineering and synthetic biology.
Contact
Dr. FENG Jian
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: jfeng@genetics.ac.cn
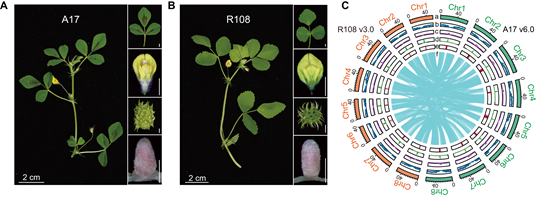 Figure. Phenotypicvariations observed between A17 and R108 (Image by IGDB)Decoding the centromere: diversity and evolutionThe researchers turned their attention to centromere, one of the most enigmatic regions of eukaryotic genomes, which plays a central role in chromosome segregation. Despite its functional importance, centromeric DNA has been historically difficult to assemble due to its highly repetitive nature. By integrating chromatin immunoprecipitation sequencing of the centromere-specific histone CENH3, fluorescence in situ hybridization (FISH), and de novo repeat annotation, the team discovered striking differences between the centromeres of A17 and R108. In A17, centromeres are primarily composed of two tandem satellite repeats: CentM168 and the species-specific CentM183. In contrast, R108 centromeres consist almost exclusively of CentM168, highlighting a distinct evolutionary trajectory between M. truncatula and M. littoralis.This divergence illustrates rapid centromere evolution, even between closely related species. The researchers propose a model in which active centromeres are maintained by CENH3-enriched CentM168 arrays, while non-active pericentromeric domains are buffered by satellite-rich flanking regions, possibly reducing chromosomal instability.In addition, they identified several highly chromosome-specific satellite repeats (e.g., CentM51, CentM515, CentM287), further underscoring the dynamic and mosaic nature of centromere evolution. Intriguingly, centromeric regions were also enriched in young long terminal repeat (LTR) retrotransposons, suggesting a potential role for transposable elements in shaping centromeric architecture and function.Implications and future directionsThe corresponding author, Dr. FENG Jian, said, “This study represents a major leap forward in our understanding of genome architecture and centromere evolution in legumes. It not only provides a high-quality genomic foundation for future gene discovery and trait mapping, but also sets the stage for engineering plant chromosomes with improved stability and transmission fidelity.”Dr. HAN Fangpu, the co-corresponding author and an expert in plant cytogenetics, added, “For decades, the centromere has been a genomic blind spot. Now, with complete assemblies and detailed repeat maps, we can begin to ask new questions about how centromeres originate, diversify, and impact legume genome evolution.”The T2T Medicago genomes generated in this work are publicly available and expected to accelerate research across legume biology, from symbiotic signaling and stress adaptation to chromosome engineering and synthetic biology.ContactDr. FENG JianInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: jfeng@genetics.ac.cn
Figure. Phenotypicvariations observed between A17 and R108 (Image by IGDB)Decoding the centromere: diversity and evolutionThe researchers turned their attention to centromere, one of the most enigmatic regions of eukaryotic genomes, which plays a central role in chromosome segregation. Despite its functional importance, centromeric DNA has been historically difficult to assemble due to its highly repetitive nature. By integrating chromatin immunoprecipitation sequencing of the centromere-specific histone CENH3, fluorescence in situ hybridization (FISH), and de novo repeat annotation, the team discovered striking differences between the centromeres of A17 and R108. In A17, centromeres are primarily composed of two tandem satellite repeats: CentM168 and the species-specific CentM183. In contrast, R108 centromeres consist almost exclusively of CentM168, highlighting a distinct evolutionary trajectory between M. truncatula and M. littoralis.This divergence illustrates rapid centromere evolution, even between closely related species. The researchers propose a model in which active centromeres are maintained by CENH3-enriched CentM168 arrays, while non-active pericentromeric domains are buffered by satellite-rich flanking regions, possibly reducing chromosomal instability.In addition, they identified several highly chromosome-specific satellite repeats (e.g., CentM51, CentM515, CentM287), further underscoring the dynamic and mosaic nature of centromere evolution. Intriguingly, centromeric regions were also enriched in young long terminal repeat (LTR) retrotransposons, suggesting a potential role for transposable elements in shaping centromeric architecture and function.Implications and future directionsThe corresponding author, Dr. FENG Jian, said, “This study represents a major leap forward in our understanding of genome architecture and centromere evolution in legumes. It not only provides a high-quality genomic foundation for future gene discovery and trait mapping, but also sets the stage for engineering plant chromosomes with improved stability and transmission fidelity.”Dr. HAN Fangpu, the co-corresponding author and an expert in plant cytogenetics, added, “For decades, the centromere has been a genomic blind spot. Now, with complete assemblies and detailed repeat maps, we can begin to ask new questions about how centromeres originate, diversify, and impact legume genome evolution.”The T2T Medicago genomes generated in this work are publicly available and expected to accelerate research across legume biology, from symbiotic signaling and stress adaptation to chromosome engineering and synthetic biology.ContactDr. FENG JianInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: jfeng@genetics.ac.cn CAS
CAS
 中文
中文




.png)
