Directed evolution—a laboratory technique that intentionally mimics natural selection—allows scientists to evolve genes and the proteins they encode. Traditionally, this technique has been used in microbes, mammalian cells, or in test tubes.
Now, however, researchers led by Prof. GAO Caixia from the Institute of Genetics and Developmental Biology (IGDB) of the Chinese Academy of Sciences (CAS) and Prof. QIU Jinlong from the Institute of Microbiology of CAS have developed a pioneering system that enables rapid and scalable directed evolution of diverse genes directly in plant cells.
The platform, called Geminivirus Replicon-Assisted in Planta Directed Evolution (
GRAPE), was published as a first release in
Science on October 2 (10.1126/scienceady2167).
Modern agricultural production requires abundant genetic resources. Directed evolution can rapidly generate genetic variants with new and enhanced properties. However, efficient platforms for performing such evolution directly in plant cells have been lacking. A key challenge is the slow cell division rate in plants, which limits the speed of selection cycles and the enrichment of functional variants.
To address this challenge, the researchers harnessed geminiviruses—plant DNA viruses that replicate DNA rapidly in plant cells via rolling circle replication (RCR). They achieved selective amplification of desired gene variants in plant cells by linking their function to the RCR of an artificial geminivirus replicon.
Building on this approach, the researchers developed GRAPE. In this platform, genes of interest (GOIs) are first mutagenized in vitro and the resulting variants are inserted into artificial geminivirus replicons. The replicon libraries are then delivered into Nicotiana benthamiana leaves, where the desired gene activity is linked to viral replication. Variants that promote replication are enriched, while those that inhibit replication are depleted. A full selection cycle can be completed on a single leaf within four days.
Using GRAPE, the researchers evolved the nucleotide-binding domain leucine-rich repeat-containing (NLR) immune receptor NRC3 to evade inhibition by the nematode effector SPRYSEC15 while preserving its immune activity. Iterative evolution of the rice NLR immune receptor Pikm-1 yielded variants that respond to six alleles of the Magnaporthe oryzae effector AVR-Pik, significantly expanding its recognition range. This strategy yields valuable genetic resources for breeding disease-resistant crops.
Compared with previous microbe-based systems, GRAPE offers distinct advantages: It excels at evolving GOIs responsible for plant-specific phenotypes (like disease resistance) or requiring plant-specific regulation, and it works directly within plant cells, eliminating the need for re-optimization. GRAPE can potentially evolve any gene functionally coupled to RCR. Beyond plant biology, GRAPE also holds promise for broader applications, such as evolving proteases to cleave specific targets for plant and pharmaceutical research.
GRAPE offers a rapid, efficient, and versatile platform that can accelerate plant synthetic biology and molecular breeding, opening new avenues for crop engineering and improving agricultural sustainability.
Figure. Graphical representation of GRAPE and its applications (Image by IGDB)
Contact:
Prof. GAO Caixia
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Email: cxgao@genetics.ac.cn
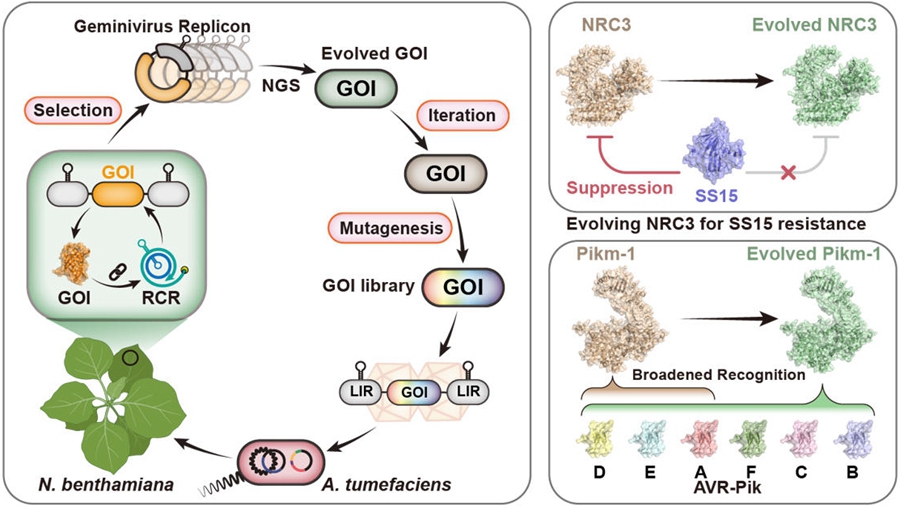 Figure. Graphical representation of GRAPE and its applications (Image by IGDB)Contact:Prof. GAO CaixiaInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: cxgao@genetics.ac.cn
Figure. Graphical representation of GRAPE and its applications (Image by IGDB)Contact:Prof. GAO CaixiaInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: cxgao@genetics.ac.cn CAS
CAS
 中文
中文




.png)
