A collaborative research team led by LU Fei from the Institute of Genetics and Developmental Biology and LIU Yongxiu from the Institute of Botany, Chinese Academy of Sciences (CAS), together with their colleagues, has unraveled the genetic basis of antagonistic selection between seed dormancy and size in wheat through a large-scale genome-wide association study.
Their findings, published in Genome Biology, offer molecular targets to boost wheat yields via precision breeding (DOI: 10.1186/s13059-025-03770-9).
Seed dormancy is a crucial adaptive mechanism in wheat, while seed size directly impacts yield and seedling vigor. During domestication and breeding, long-term human selection for “large kernel” and “easy germination” traits has resulted in modern cultivars with generally larger seeds but reduced dormancy, increasing the risk of pre-harvest sprouting and leading to significant yield losses. Unraveling the genetic relationship between these two traits and breaking the “large kernel-weak dormancy” linkage is a key challenge in modern breeding.
The team conducted a genome-wide association study (GWAS) on 545 representative wheat accessions. They systematically revealed the genome-wide genetic basis underlying the antagonism between seed dormancy and size. The study found significant antagonism between variations associated with the two traits: 18.59% to 28.60% of significant markers carried antagonistic alleles, while only 0.21% to 0.76% were located at synergistic loci. Further analysis showed that during breeding, superior alleles controlling larger kernels were consistently selected, while alleles enhancing dormancy significantly decreased in frequency. This has led to an increasing abundance of "large kernel" genes and a declining presence of "strong dormancy" genes in modern cultivars, creating a significant genetic trade-off. This trend has become more pronounced with advancing breeding cycles, resulting in a significant negative genetic correlation between the two traits.
The research focused on elucidating two primary genetic mechanisms responsible for this trait antagonism: pleiotropy and antagonistic linkage. In terms of pleiotropy, for example, Tamyb10not only enhances seed dormancy and induces red pigmentation in the seed coat but also suppresses seed enlargement. TaGW2, on the other hand, negatively regulates seed size while positively regulating dormancy; its overexpression results in smaller seeds and deeper dormancy. Regarding antagonistic linkage, genome-wide analysis identified 66 genomic regions concurrently associated with both seed size and dormancy, 21 of which exhibited significant antagonistic effects. Within these regions, genes controlling "large kernel" and "weak dormancy" are tightly linked, forming “gene blocks” that are difficult to break through conventional breeding, thereby complicating coordinated improvement.
The study also identified three overlapping regions with synergistic potential for improvement. From one such region, a key candidate gene, TaRBP-4A, was mined. Its GG haplotype was found to simultaneously enhance dormancy and increase kernel weight, demonstrating excellent breeding potential.
This research is the first to reveal the genome-wide genetic architecture of antagonism between seed size and dormancy in wheat. It identifies multiple key genes and rare synergistic loci and develops corresponding molecular markers, providing new strategies and genetic resources for breaking trait trade-offs and achieving coordinated improvement.
This study, a collaboration across multiple Chinese institutions, was supported by the National Key Research and Development Program of China, the CAS Strategic Priority Research Program, and the National Natural Science Foundation of China.
Figure 1. Tamyb10 and TaGW2 affect both seed dormancy and seed size in wheat (Image by IGDB)
Contacts:
Dr. LU Fei
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
Dr. LIU Yongxiu
Institute of Botany, Chinese Academy of Sciences
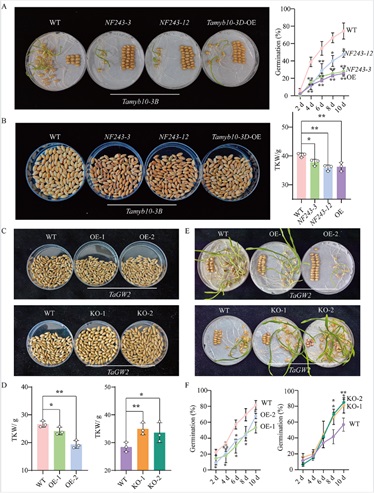 Figure 1. Tamyb10 and TaGW2 affect both seed dormancy and seed size in wheat (Image by IGDB)Contacts:Dr. LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: flu@genetics.ac.cnDr. LIU YongxiuInstitute of Botany, Chinese Academy of SciencesEmail: yongxiu@ibcas.ac.cn
Figure 1. Tamyb10 and TaGW2 affect both seed dormancy and seed size in wheat (Image by IGDB)Contacts:Dr. LU FeiInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: flu@genetics.ac.cnDr. LIU YongxiuInstitute of Botany, Chinese Academy of SciencesEmail: yongxiu@ibcas.ac.cn CAS
CAS
 中文
中文




.png)
