-
Scientists Develop “Mortise-Tenon Joint System” to Advance Precise Rice Genome Editing
TIME: 26 Nov 2025Precise and scarless DNA insertion and replacement represent a core challenge in plant genome editing, with profound significance for crop breeding technologies and food security. The current mainstream precision editing technologies are predominantly based on the prime editing (PE) system. However, the concentrated ownership of their core patents restricts their widespread and flexible application in academic research and industrial practice.On November 18, 2025, researchers led by ZHANG Huawei from Peking University and LI Jiayang from the Institute of Genetics and Developmental Biology of the Chinese Academy of Sciences/ Yazhouwan National Laboratory published an innovative study in Molecular Plant (https://doi.org/10.1016/j.molp.2025.11.006) titled "A Mortise-Tenon Joint System Facilitates Precise Targeted DNA Insertion and Replacement in Rice." The study developed a novel genome editing strategy: the "Mortise-Tenon system" (MT). This system achieved precise insertion and replacement efficiencies of 16.30%–59.47% in rice, providing a new tool for plant genome editing and opening new avenues for crop genetic improvement.Inspired by the traditional woodworking mortise-tenon structure, the MT system's core lies in the precise complementary pairing of the "mortise" and "tenon." The research team utilized the APOBEC-Cas9-UDG/AP endonuclease complex from their previously developed AFID system to generate unique double-strand break structures at the target genomic site, such as "mortises" with single-stranded or double-stranded 5' overhangs.Simultaneously, they designed double-stranded DNA donors with complementary 5' sticky ends as "tenons," enabling precise insertion or replacement of donor fragments through end-capture. The MT system boasts three notable advantages: First, high specificity. The MT2 subsystem leverages APOBEC3B's specificity for TC motifs to generate precisely sized sticky ends, effectively avoiding off-target editing. Second, broad applicability. It can achieve efficient editing for both single and multiple TC motif target sites by designing corresponding sticky-ended donors. Third, comprehensive functionality. It enables precise insertion of 21–85 bp fragments, as well as fragment replacement, with heritable editing events.Tests at multiple rice gene loci (e.g., GRF1, NRT1.1B, IPA1) demonstrated that the MT system's editing efficiency significantly outperformed traditional Cas9-based methods, reaching up to 59.47%—over 10 times higher than that of conventional Cas9-mediated editor.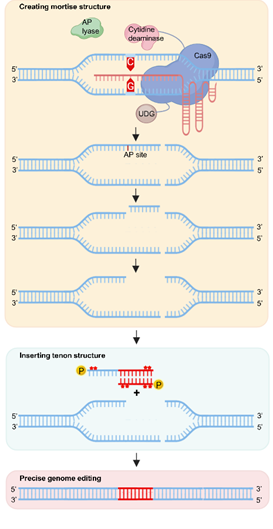 Schematic Diagram of MT System (Image by SUN et al.)Contact:Prof. LI JiayangInstitute of Genetics and Developmental Biology, Chinese Academy of Sciences
Schematic Diagram of MT System (Image by SUN et al.)Contact:Prof. LI JiayangInstitute of Genetics and Developmental Biology, Chinese Academy of SciencesEmail: jyli@genetics.ac.cn
 CAS
CAS
 中文
中文




.png)
