PUBLICATIONS (# first author; * corresponding author)
Zhang L#, Gao Y#, Xu Z, Tian Y, Hu J, Wen Z, Li J, Gao C, Zhou Y*, Zhang B*. (2025) Shaping pit structure in vessel walls sustains xylem hydraulics and grain yield. Cell 188: 7238–7251.e15.
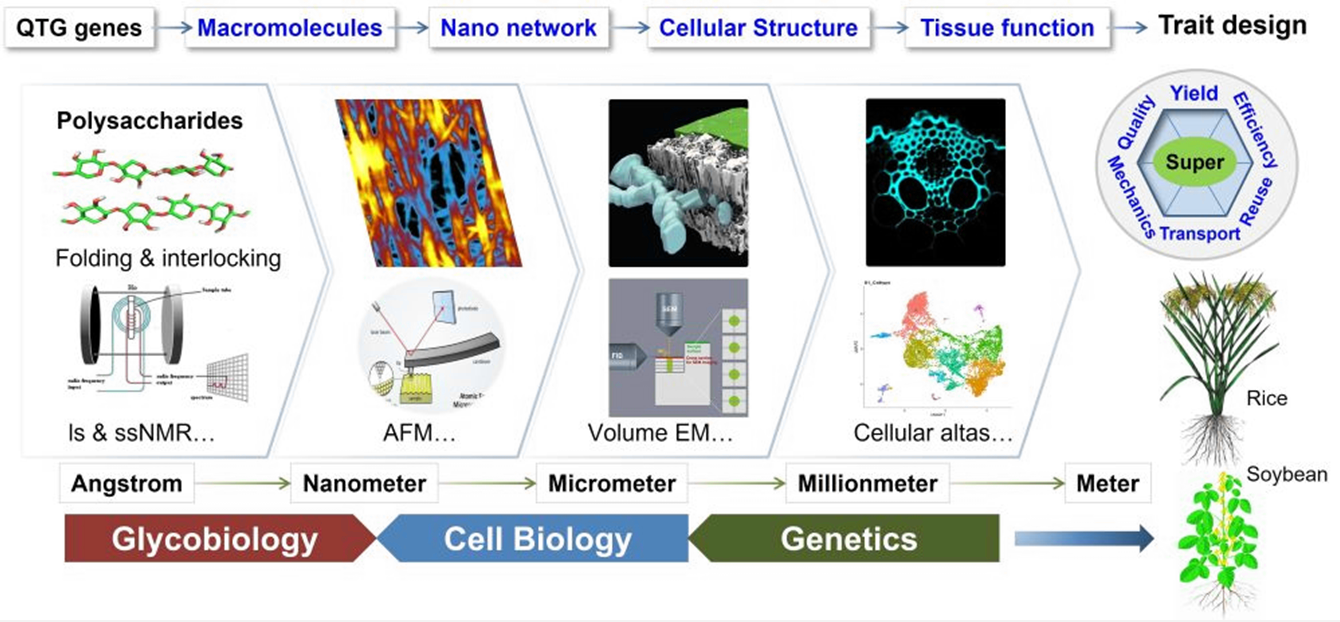
Zhang L, Gao C, Gao Y, Yang H, Jia M, Wang X, Zhang B*, Zhou Y*. (2025) New insights into plant cell wall functions. J Genet Genomics 52: 1308–1324.
Wen Z#, Xu Z#, Zhang L#, Xue Y#, Wang H, Jian L, Ma J, Liu Z, Yang H, Huang S, Kang X, Zhou Y, Zhang B*. (2025) XYLAN O-ACETYLTRANSFERASE 6 promotes xylan synthesis by forming a complex with IRX10 and governs wall formation in rice. Plant Cell 37: koae322.
Zhou Y#, Gao Y#, Zhang B#, Yang H, Tian Y, Huang Y, Yin C, Tao J, Wei W, Zhang W, Chen S, Zhou Y*, Zhang J*. (2024). CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice. Plant Cell 36: 3751–3769.
Zhang L, Zhou Y, Zhang B*. 2024. Xylan-directed cell wall assembly in grasses. Plant Physiol 194: 2197–2207. (Invited review)
Xu R#, Liu Z#, Zhou Y, Zhang B*. (2024) Xylan clustering on the pollen surface is required for exine patterning. Plant Physiol 194: 153-167. (Invited research article)
Cao S, Wang Y, Gao Y, Xu R, Ma J, Xu Z, Shang-Guan K, Zhang B*, Zhou Y* (2023) The RLCK-VND6 module coordinates secondary cell wall formation and adaptive growth in rice. Mol Plant 16: 999-1015.
Xu Z#, Gao Y#, Gao C#, Mei J, Wang S, Ma J, Yang H, Cao S, Wang Y, Zhang F, Liu X, Liu Q, Zhou Y*, Zhang B*. (2022) Glycosylphosphatidylinositol anchor lipid remodeling directs proteins to the plasma membrane and governs cell wall mechanics. Plant Cell 34: 4778–4794.
(Highlighted by The Plant Cell 2022, 34: 4671–4672)
Wang H#, Yang H#, Wen Z, Gao C, Gao Y, Tian Y, Xu Z, Liu X, Persson S, Zhang B*, Zhou Y*. (2022) Xylan-based nanocompartments orchestrate plant vessel wall patterning. Nat Plants 8: 295–306.
(Highlighted by Research Briefing in Nature Plants 2022, 8:330–331)
Zhou Y, Zhang B. (2022) Unprecedented polysaccharide nanostructures sustain vessel wall patterning and robustness. Nat Plants 8: 330–331. (Research Briefing)
Peng J#, Zhang B#, Chen H#, Wang M#, Wang Y, Li H, Cao S, Yi H, Wang H, Zhou Y*, Gong J*. (2021). Galactosylation of rhamnogalacturonan-II for cell wall pectin biosynthesis is critical for root apoplastic iron reallocation in Arabidopsis. Mol Plant 14: 1640–1651.
Zhang B, Gao Y, Zhang L, Zhou Y*. (2021) The plant cell wall: biosynthesis, construction, and functions. J Integr Plant Biol 63: 251–272.
Cai Y#, Zhang B#, Liang L#, Wang S, Zhang L, Wang L, Cui H*, Zhou Y*, Wang D*. (2021) A solid-state nanopore-based single-molecule approach for label-free characterization of plant polysaccharides. Plant Commun 2: 100106.
Gao Y#, Xu Z#, Zhang L#, Li S, Wang S, Yang H, Liu X, Zeng D, Liu Q, Qian Q, Zhang B*, Zhou Y*. (2020) MYB61 is regulated by GRF4 and promotes nitrogen utilization and biomass production in rice. Nat Commun 11: 5219.
Xue J#, Zhang B#, Zhan H#, Lv Y, Jia X, Wang T, Yang N, Lou Y, Zhang Z, Hu W, Gui J, Cao J, Xu P, Zhou Y, Hu J, Li L*, Yang Z*. (2020) Phenylpropanoid derivatives are essential components of sporopollenin in vascular plants. Mol Plant 13: 1644–1653.
Zhang L, Zhang B*, Zhou Y*. (2019) Cell wall compositional analysis of rice culms. Bio-protocol 9: e3398.
Wang S#, Yang H#, Mei J, Liu X, Wen Z, Zhang L, Xu Z, Zhang B*, Zhou Y*. (2019) A rice homeobox protein KNAT7 integrates the pathways regulating cell expansion and wall stiffness. Plant Physiol 181: 669–682.
(Highlighted with News and Views in Plant Physiol 181(2): 385-386)
Zhang L, Gao C, Mentink-Vigier F, Tang L, Zhang D, Wang S, Cao S, Xu Z, Liu X, Wang T, Zhou Y*, Zhang B*. (2019) Arabinosyl deacetylase modulates the arabinoxylan acetylation profile and secondary wall formation. Plant Cell 31: 1113–1126.
(Highlighted with In Brief in Plant Cell 31(5):936)
Zhang Z
#,
ZhangB#, Chen Z, Zhang D,
Zhang
H, Wang H, Zhang Y, Cai D, Liu J, Xiao S, Huo Y, Liu Jie, Zhang L, Wang M, Liu X,
Xue Y, Zhao L*, Zhou Y*, Chen H*. (2018) A
PECTIN METHYLESTERASE at the Maize
Ga1 locus confers male function in unilateral cross-incompatibility.
Nat Commun 9: 3678.
Zhang D, Xu Z, Cao S, Chen K, Li S, Liu X, Gao C, Zhang B*, Zhou Y*. (2018) An uncanonical CCCH-tandem zinc finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol Plant 11: 163–174.
Zhang B#, Zhang L#, Li F#, Zhang M, Liu X, Wang H, Xu Z, Chu C*, and Zhou Y*. (2017) Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat Plants 3: 17017.
(Highlighted with News and Views in Nat Plants 3: 17024)
Gao Y#, He C#, Zhang D, Liu X, Xu Z, Tian Y, Liu XH, Zang S, Pauly M, Zhou Y*, Zhang B*. (2017) Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol 173: 470–481.
Zhang B and Zhou Y. (2017) Carbohydrate composition analysis in xylem. Methods Mol Biol 1544: 213–222. (Chapter)
Shi Y, Liu X, Li R, Gao Y, Xu Z, Zhang B*, Zhou Y. Retention of OsNMD3 in the cytoplasm disturbs protein synthesis efficiency and affects plant development in rice. (2014) J Exp Bot 65: 3055–3069.
Wu B#, Zhang B#, Dai Y#, Zhang L, Shang-Guan K, Peng Y, Zhou Y*, Zhu Z*. (2012) Brittle Culm15 encodes a membrane-associated chitinase-like protein required for cellulose biosynthesis in rice. Plant Physiol 159: 1440–1452.
Ning J#, Zhang B#, Wang N, Zhou Y*, and Xiong L*. (2011) Increased leaf angle1, a Raf-Like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the lamina joint of rice. Plant Cell 23: 4334–4347.
Zhang B#, Liu X#, Qian Q#, Liu L, Dong G, Xiong G, Zeng D, and Zhou Y. (2011) A Golgi nucleotide sugar transporter modulates cell wall biosynthesis and plant growth in rice. Proc Natl Acad Sci USA 108: 5110–5115.
Zhang B and Zhou Y. (2011) Study on rice brittleness mutants: A way to open the ‘black box’ of monocot cell wall biosynthesis. J Integr Plant Biol 53: 136–142.
Zhang M#, Zhang B#, Qian Q#, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y. (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63: 312–328.
Zhang B#, Deng L#, Qian Q#, Xiong G, Zeng D, Li R, Guo L, Li J, Zhou Y. (2009) A missense mutation in the transmembrane domain of CESA4 affects protein abundance in the plasma membrane and results in abnormal cell wall biosynthesis in rice. Plant Mol Biol 71: 509–524.
Updated at Jan 2026
(Full publication list can be found in https://www.researchgate.net/profile/Baocai-Zhang/research)


 CAS
CAS
 中文
中文




.png)