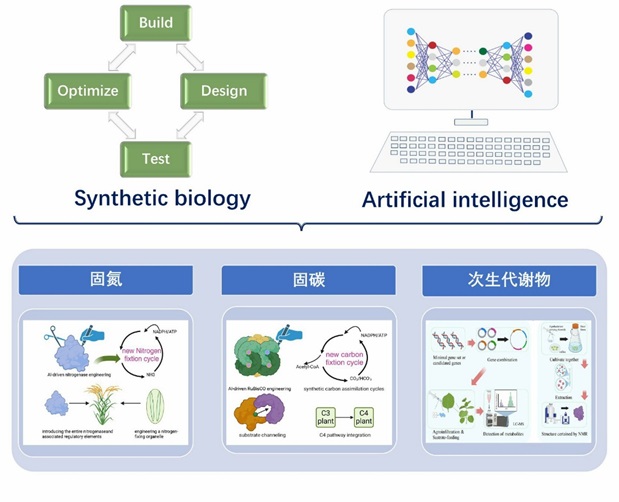
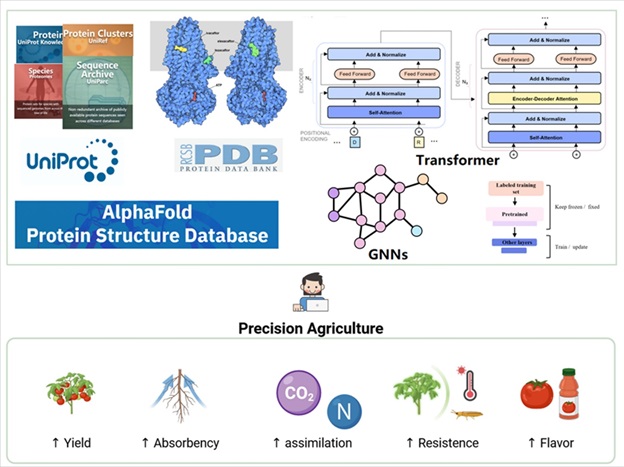
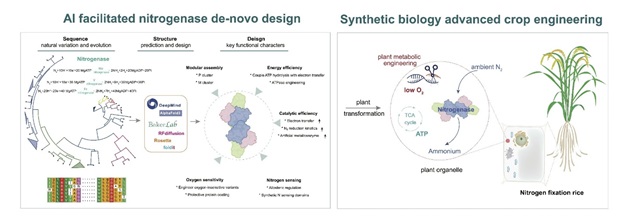
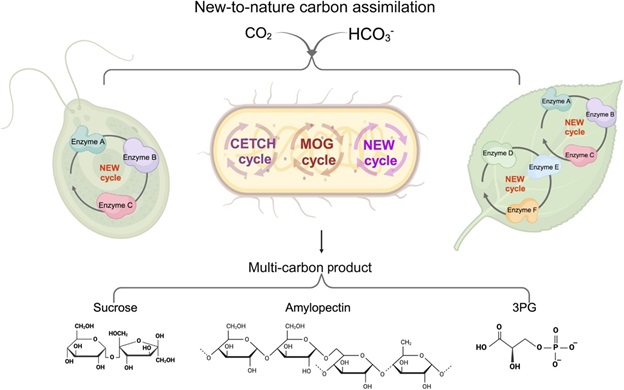
Representative Works:
Publications:(#Co-first author, *corresponding author)
86. Wu S., Zhang Y., Luzarowska U., Yang L., Salem M. A., Thirumalaikumar V. P., Sade N., Galperin V. E., Fernie A., Sampathkumar A., Bershtein S., Fusari C. M. & Brotman Y. (2025). The Homeostasis of Beta-Alanine Is Key for Arabidopsis Reproductive Growth and Development. The Plant Journal 122(1): e70134.
85. Qin K., Ye X., Luo S., Fernie A.R. & Zhang Y. (2025). Engineering Carbon Assimilation in Plants. Journal of Integrative Plant Biology 67(4): 926-948.
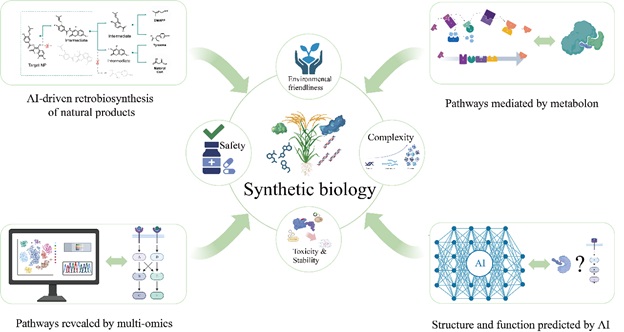
84. Qin K., Liu F., Zhang C., Deng R., Fernie A.R. & Zhang Y. (2025). Systems and Synthetic Biology for Plant Natural Product Pathway Elucidation. Cell Reports 44(6): 115715.
83. Nejamkin A., Decima Oneto C.,
Zhang Y., Luquet M., Galatro A., Del Castello F., Massa G.A., Feingold S., Fernie A.R., Lamattina L., Correa-Aragunde N.& Foresi N. (2025).
Metabolic Profiling Reveals Increased Nitrogen Remobilization in Potato Plants That Express a Cyanobacterial No/No3-Producing Enzyme. Journal of Plant Growth Regulation
82.
Liu F.,
Zhao Z., Fernie A.R. &
Zhang Y. (2025).
Towards establishing functional nitrogenase activities within plants. Trends in Biotechnology
81. Decros G., Zhang Y. & Fernie A.R. (2025). Mitochondrial Support of High Rates of Photosynthesis. Plant Communications 6(3): 101285.
80. Cao Y., Chen Q., Xu X., Fernie A.R., Li J. & Zhang Y. (2025). Insights from Natural Rubber Biosynthesis Evolution for Pathway Engineering. Trends in Plant Science
79. Zhang Y., Qin K. & Fernie A.R. (2024). Plant Tissue Culture and Metabolite Profiling for High-Value Natural Product Synthesis. Methods in Molecular Biology 2827: 405-416.
77. Timm S., Klaas N., Niemann J., Jahnke K., Alseekh S., Zhang Y., Souza P. V. L., Hou L. Y., Cosse M., Selinski J., Geigenberger P., Daloso D. M., Fernie A.R. & Hagemann M. (2024). Thioredoxins O1 and H2 Jointly Adjust Mitochondrial Dihydrolipoamide Dehydrogenase-Dependent Pathways Towards Changing Environments. Plant Cell & Environment 47(7): 2542-2560.
75. Luo M., Yang W., Bai L., Zhang L., Huang J.W., Cao Y., Xie Y., Tong L., Zhang H., Yu L., Zhou L.W., Shi Y., Yu P., Wang Z., Yuan Z., Zhang P., Zhang Y., Ju F., Zhang H., Wang F., Cui Y., Zhang J., Jia G., Wan D., Ruan C., Zeng Y., Wu P., Gao Z., Zhao W., Xu Y., Yu G., Tian C., Jin L.N., Dai J., Xia B., Sun B., Chen F., Gao Y.Z, Wang H., Wang B., Zhang D., Cao X., Wang H. &Huang T. (2024). Artificial Intelligence for Life Sciences: A Comprehensive Guide and Future Trends. The Innovation Life 2(4): 100105.
74. Liu F., Fernie A.R. & Zhang Y. (2024). Can a Nitrogen-Fixing Organelle Be Engineered within Plants? Trends in Plant Science 29(11): 1168-1171.
73. Liu F., Fernie A.R. & Zhang Y. (2024). Plant Gene Co-Expression Defines the Biosynthetic Pathway of Neuroactive Alkaloids. Molecular Plant 17(3): 372-374.
72. Fernie A.R., Liu F. & Zhang Y. (2024). Post-Genomic Illumination of Paclitaxel Biosynthesis. Nature Plants 10(12): 1875-1885.
71. Dorrell R.G., Zhang Y., Liang Y., Gueguen N., Nonoyama T., Croteau D., Penot-Raquin M., Adiba S., Bailleul B., Gros V., Pierella Karlusich J.J., Zweig N., Fernie A.R., Jouhet J., Marechal E. & Bowler C. (2024). Complementary Environmental Analysis and Functional Characterization of Lower Glycolysis-Gluconeogenesis in the Diatom Plastid. The Plant Cell 36(9): 3584-3610.
70. Zhang Y., Wiese L., Fang H., Alseekh S., de Souza L.P., Scossa F., Molloy J.J., Christmann M. & Fernie A.R. (2023). Synthetic Biology Identifies the Minimal Gene Set Required for Paclitaxel Biosynthesis in a Plant Chassis. Molecular Plant 16(12): 1951-1961.
69. Zhang Y., Jaime S. M., Bulut M., Graf A. & Fernie A.R. (2023). The Conditional Mitochondrial Protein Complexome in the Arabidopsis Thaliana Root and Shoot. Plant Communications 4(5): 100635.
68. Zhang Y. & Fernie A.R. (2023). The Role of TCA Cycle Enzymes in Plants. Advanced Biology 7(8): e2200238.
67. Zhang Y. & Fernie A.R. (2023). Leveraging Glycoside-Targeted Metabolomics to Gain Insight into Biological Function. Trends in Plant Science 28(7): 737-739.
66.
Zhang Y., Chen M., Liu T.,
Qin K. & Fernie A.R. (2023). Investigating the Dynamics of Protein-Protein Interactions in Plants.
The Plant Journal 114(4): 965-983.
65. Xie J.Q., Zhou X., Jia Z.C., Su C.F., Zhang Y., Fernie A.R., Zhang J., Du Z. Y. & Chen M.X. (2023). Alternative Splicing, an Overlooked Defense Frontier of Plants with Respect to Bacterial Infection. Journal of Agricultural and Food Chemistry 71(45): 16883-16901.
64. Wan H., Zhang Y., Wu L., Zhou G., Pan L., Fernie A.R. & Ruan Y.L. (2023). Evolution of Cytosolic and Organellar Invertases Empowered the Colonization and Thriving of Land Plants. Plant Physiology 193(2): 1227-1243.
63. Su C., Kokosza A., Xie X., Pencik A., Zhang Y., Raumonen P., Shi X., Muranen S., Topcu M.K., Immanen J., Hagqvist R., Safronov O., Alonso-Serra J., Eswaran G., Venegas M.P., Ljung K., Ward S., Mahonen A.P., Himanen K., Salojarvi J., Fernie A.R., Novak O., Leyser O., Palubicki W., Helariutta Y. & Nieminen K. (2023). Tree Architecture: A Strigolactone-Deficient Mutant Reveals a Connection between Branching Order and Auxin Gradient Along the Tree Stem. Proceedings of the National Academy of Sciences 120(48): e2308587120.
62. Song Y.C., Das D., Zhang Y., Chen M.X., Fernie A.R., Zhu F.Y. & Han J. (2023). Proteogenomics-Based Functional Genome Research: Approaches, Applications, and Perspectives in Plants. Trends in Biotechnology 41(12): 1532-1548.
61. Siqueira J. A., Zhang Y., Nunes-Nesi A., Fernie A.R. & Araujo W. L. (2023). Beyond Photorespiration: The Significance of Glycine and Serine in Leaf Metabolism. Trends in Plant Science 28(10): 1092-1094.
60. Shan N., Zhang Y., Guo Y., Zhang W., Nie J., Fernie A.R. & Sui X. (2023). Cucumber Malate Decarboxylase, Csnadp-Me2, Functions in the Balance of Carbon and Amino Acid Metabolism in Fruit. Horticulture Research 10(12): uhad216.
59. Schlossarek D., Zhang Y., Sokolowska E. M., Fernie A.R., Luzarowski M. & Skirycz A. (2023). Don't Let Go: Co-Fractionation Mass Spectrometry for Untargeted Mapping of Protein-Metabolite Interactomes. The Plant Journal 113(5): 904-914.
58. Natale R., Coppola M., D'Agostino N., Zhang Y., Fernie A.R., Castaldi V. & Rao R. (2023). In Silico and in Vitro Approaches Allow the Identification of the Prosystemin Molecular Network. Computational and Structural Biotechnology Journal 21: 212-223.
57. Jia Z.C., Das D., Zhang Y., Fernie A.R., Liu Y.G., Chen M. & Zhang J. (2023). Plant Serine/Arginine-Rich Proteins: Versatile Players in Rna Processing. Planta 257(6): 109.
56. Giese J., Eirich J., Walther D., Zhang Y., Lassowskat I., Fernie A.R., Elsasser M., Maurino V.G., Schwarzlander M. & Finkemeier I. (2023). The Interplay of Post-Translational Protein Modifications in Arabidopsis Leaves During Photosynthesis Induction. The Plant Journal 116(4): 1172-1193.
55. Fernie A.R. & Zhang Y. (2023). Observations and Implication of Thermal Tolerance in the Arabidopsis Proteome. Cell Systems 14(10): 819-821.
54. Dahmani I., Qin K., Zhang Y. & Fernie A.R. (2023). The Formation and Function of Plant Metabolons. The Plant Journal 114(5): 1080-1092.
53. Chen Y., Wang Y., Liang X., Zhang Y. & Fernie A.R. (2023). Mass Spectrometric Exploration of Phytohormone Profiles and Signaling Networks. Trends in Plant Science 28(4): 399-414.
52. Bao Y.J., Chen J.X., Zhang Y., Fernie A.R., Zhang J., Huang B.X., Zhu F.Y. & Cao F.L. (2023). Emerging Role of Jasmonic Acid in Woody Plant Development. Advanced Agrochem 3(1): 26-38.
51. Zhang Y. & Fernie A.R. (2022). Dynamically Regulating Metabolic Fluxes with Synthetic Metabolons. Trends in Biotechnology 40(9): 1019-1020.
50. Zhang Y. & Fernie A.R. (2022). Metabolite Profiling of Arabidopsis Mutants of Lower Glycolysis. Scientific Data 9(1): 614.
49. Stefan T., Wu X.N., Zhang Y., Fernie A. & Schulze W.X. (2022). Regulatory Modules of Metabolites and Protein Phosphorylation in Arabidopsis Genotypes with Altered Sucrose Allocation. Frontiers in Plant Science 13: 891405.
48. Fernie A.R. & Zhang Y. (2022). The Bacillus Subtilis Glutamate Anti-Metabolon. Nature Metabolism 4(2): 161-162.
47. Amanda D., Frey F.P., Neumann U., Przybyl M., Simura J., Zhang Y., Chen Z., Gallavotti A., Fernie A.R., Ljung K. & Acosta I. F. (2022). Auxin Boosts Energy Generation Pathways to Fuel Pollen Maturation in Barley. Current Biology 32(8): 1798-1811.
46. Zhang Y., Scossa F. & Fernie A.R. (2021). The Genomes of Taxus Species Unveil Novel Candidates in the Biosynthesis of Taxoids. Molecular Plant 14(11): 1773-1775.
45. Zhang Y., Giese J., Kerbler S.M., Siemiatkowska B., de Souza L.P., Alpers J., Medeiros D.B., Hincha D.K., Daloso D.M., Stitt M., Finkemeier I. & Fernie A.R. (2021). Two Mitochondrial Phosphatases, PP2c63 and Sal2, Are Required for Posttranslational Regulation of the TCA Cycle in Arabidopsis. Molecular Plant 14(7): 1104-1118.
44. Zhang Y. & Fernie A.R. (2021). Stable and Temporary Enzyme Complexes and Metabolons Involved in Energy and Redox Metabolism. Antioxidants & Redox Signaling 35(10): 788-807.
43. Zhang Y. & Fernie A.R. (2021). Metabolons, Enzyme-Enzyme Assemblies That Mediate Substrate Channeling, and Their Roles in Plant Metabolism. Plant Communications 2(1): 100081.
42. Wu T., Kerbler S.M., Fernie A.R. & Zhang Y. (2021). Plant Cell Cultures as Heterologous Bio-Factories for Secondary Metabolite Production. Plant Communications 2(5): 100235.
41. Qin K., Fernie A.R. & Zhang Y. (2021). The Assembly of Super-Complexes in the Plant Chloroplast. Biomolecules 11(12): 1839.
40. Pereira L., Sapkota M., Alonge M., Zheng Y., Zhang Y., Razifard H., Taitano N.K., Schatz M.C., Fernie A.R., Wang Y., Fei Z., Caicedo A.L., Tieman D.M. & van der Knaap E. (2021). Natural Genetic Diversity in Tomato Flavor Genes. Frontiers in Plant Science 12: 642828.
39. Moreno J.C., Rojas B.E., Vicente R., Gorka M., Matz T., Chodasiewicz M., Peralta-Ariza J.S., Zhang Y., Alseekh S., Childs D., Luzarowski M., Nikoloski Z., Zarivach R., Walther D., Hartman M.D., Figueroa C.M., Iglesias A.A., Fernie A.R. & Skirycz A. (2021). Tyr-Asp Inhibition of Glyceraldehyde 3-Phosphate Dehydrogenase Affects Plant Redox Metabolism. EMBO Journal 40(15): e106800.
38. Kerbler S. M., Natale R., Fernie A.R. & Zhang Y. (2021). From Affinity to Proximity Techniques to Investigate Protein Complexes in Plants. International Journal of Molecular Sciences 22(13): 7101.
37. Chen M.X., Zhang Y., Fernie A.R., Liu Y.G. & Zhu F.Y. (2021). Swath-Ms-Based Proteomics: Strategies and Applications in Plants. Trends in Biotechnology 39(5): 433-437.
36. Cervela-Cardona L., Yoshida T., Zhang Y., Okada M., Fernie A. & Mas P. (2021). Circadian Control of Metabolism by the Clock Component Toc1. Frontiers in Plant Science 12: 683516.
35. Aarabi F., Rakpenthai A., Barahimipour R., Gorka M., Alseekh S., Zhang Y., Salem M.A., Bruckner F., Omranian N., Watanabe M., Nikoloski Z., Giavalisco P., Tohge T., Graf A., Fernie A.R. & Hoefgen R. (2021). Sulfur Deficiency-Induced Genes Affect Seed Protein Accumulation and Composition under Sulfate Deprivation. Plant Physiology 187(4): 2419-2434.
34. Zhu J., Loubéry S., Broger L., Zhang Y., Lorenzo-Orts L., Utz-Pugin A., Fernie A.R., Young-Tae C. & Hothorn M. (2020). A Genetically Validated Approach for Detecting Inorganic Polyphosphates in Plants. The Plant Journal 102(3): 507-516.
33. Zhang Y., Skirycz A. & Fernie A.R. (2020). An Abundance and Interaction Encyclopedia of Plant Protein Function. Trends in Plant Science 25(7): 627-630.
32. Zhang Y., Sampathkumar A., Kerber S.M., Swart C., Hille C., Seerangan K., Graf A., Sweetlove L. & Fernie A.R. (2020). A Moonlighting Role for Enzymes of Glycolysis in the Co-Localization of Mitochondria and Chloroplasts. Nature Communications 11(1): 4509.
29. Zhang Y. & Fernie A.R. (2020). Resolving the Metabolon: Is the Proof in the Metabolite? EMBO Reports 21(8): e50774.
28. Zhang Y., Chen M., Siemiatkowska B., Toleco M.R., Jing Y., Strotmann V., Zhang J., Stahl Y. & Fernie A.R. (2020). A Highly Efficient Agrobacterium-Mediated Method for Transient Gene Expression and Functional Studies in Multiple Plant Species. Plant Communications 1(5): 100028.
27. Zhang W., Zhang Y., Qiu H., Guo Y., Wan H., Zhang X., Scossa F., Alseekh S., Zhang Q., Wang P., Xu L., Schmidt M.H., Jia X., Li D., Zhu A., Guo F., Chen W., Ni D., Usadel B., Fernie A.R. & Wen W. (2020). Genome Assembly of Wild Tea Tree Dasz Reveals Pedigree and Selection History of Tea Varieties. Nature Communications 11(1): 3719.
25. Tohge T., Scossa F., Wendenburg R., Frasse P., Balbo I., Watanabe M., Alseekh S., Jadhav S.S., Delfin J.C., Lohse M., Giavalisco P., Usadel B., Zhang Y., Luo J., Bouzayen M. & Fernie A.R. (2020). Exploiting Natural Variation in Tomato to Define Pathway Structure and Metabolic Regulation of Fruit Polyphenolics in the Lycopersicum Complex. Molecular Plant 13(7): 1027-1046.
24. Fernie A.R., Zhang Y. & Sampathkumar A. (2020). Cytoskeleton Architecture Regulates Glycolysis Coupling Cellular Metabolism to Mechanical Cues. Trends in Biochemical Science 45(8): 637-638.
23. Chen M.X., Zhu F.Y., Gao B., Ma K.L., Zhang Y., Fernie A.R., Chen X., Dai L., Ye N.H., Zhang X., Tian Y., Zhang D., Xiao S., Zhang J. & Liu Y.G. (2020). Full-Length Transcript-Based Proteogenomics of Rice Improves Its Genome and Proteome Annotation. Plant Physiology 182(3): 1510-1526.
22. Zhu J., Lau K., Puschmann R., Harmel R.K., Zhang Y., Pries V., Gaugler P., Broger L., Dutta A.K., Jessen H.J., Schaaf G., Fernie A.R., Hothorn L.A., Fiedler D. & Hothorn M. (2019). Two Bifunctional Inositol Pyrophosphate Kinases/Phosphatases Control Plant Phosphate Homeostasis. Elife 8: e43582.
21. Zhang Y., Natale R., Domingues A.P. Junior, Toleco M.R., Siemiatkowska B., Fabregas N. & Fernie A.R. (2019). Rapid Identification of Protein-Protein Interactions in Plants. Current Protocals in Plant Biology 4(4): e20099.
20. Reinholdt O., Schwab S., Zhang Y., Reichheld J.P., Fernie A.R., Hagemann M. & Timm S. (2019). Redox-Regulation of Photorespiration through Mitochondrial Thioredoxin O1. Plant Physiology 181(2): 442-457.
19. Liu T.Y., Chen M.X., Zhang Y., Zhu F.Y., Liu Y.G., Tian Y., Fernie A.R., Ye N. & Zhang J. (2019). Comparative Metabolite Profiling of Two Switchgrass Ecotypes Reveals Differences in Drought Stress Responses and Rhizosheath Weight. Planta 250(4): 1355-1369.
18. Zhang Y., Swart C., Alseekh S., Scossa F., Jiang L., Obata T., Graf A. & Fernie A.R. (2018). The Extra-Pathway Interactome of the TCA Cycle: Expected and Unexpected Metabolic Interactions. Plant Physiology 177(3): 966-979.
17. Zhang Y., Zhang Y., McFarlane H.E., Obata T., Richter A.S., Lohse M., Grimm B., Persson S., Fernie A.R. & Giavalisco P. (2018). Inhibition of TOR Represses Nutrient Consumption, Which Improves Greening after Extended Periods of Etiolation. Plant Physiology 178(1): 101-117.
16. Zhang Y., Giuliani R., Zhang Y., Zhang Y., Araujo W.L., Wang B., Liu P., Sun Q., Cousins A., Edwards G., Fernie A., Brutnell T.P. & Li P. (2018). Characterization of Maize Leaf Pyruvate Orthophosphate Dikinase Using High Throughput Sequencing. Journal of integrative plant biology 60(8): 670-690.
15. Zhang Y. & Fernie A.R. (2018). On the Role of the Tricarboxylic Acid Cycle in Plant Productivity. Journal of Integrative Plant Biology 60(12): 1199-1216.
14. Scossa F., Benina M., Alseekh S., Zhang Y. & Fernie A.R. (2018). The Integration of Metabolomics and Next-Generation Sequencing Data to Elucidate the Pathways of Natural Product Metabolism in Medicinal Plants. Planta Medica 84(12-13): 855-873.
13. Ma X.,
Zhang Y., Tureckova V., Xue G.P., Fernie A.R., Mueller-Roeber B. & Balazadeh S. (2018). The NAC Transcription Factor
SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiology 177(3): 1286-1302.
11. Andersson M., Turesson H., Arrivault S., Zhang Y., Falt A.S., Fernie A.R. & Hofvander P. (2018). Inhibition of Plastid Ppase and Ntt Leads to Major Changes in Starch and Tuber Formation in Potato. Journal of Experimental Botany 69(8): 1913-1924.
10. Zhu F.Y., Chen M.X., Ye N.H., Shi L., Ma K.L., Yang J.F., Cao Y.Y., Zhang Y., Yoshida T., Fernie A.R., Fan G.Y., Wen B., Zhou R., Liu T.Y., Fan T., Gao B., Zhang D., Hao G.F., Xiao S., Liu Y.G. & Zhang J. (2017). Proteogenomic Analysis Reveals Alternative Splicing and Translation as Part of the Abscisic Acid Response in Arabidopsis Seedlings. The Plant Journal 91(3): 518-533.
9. Zhang Y., Beard K.F.M., Swart C., Bergmann S., Krahnert I., Nikoloski Z., Graf A., Ratcliffe R.G., Sweetlove L.J., Fernie A.R. & Obata T. (2017). Protein-Protein Interactions and Metabolite Channelling in the Plant Tricarboxylic Acid Cycle. Nature Communications 8: 15212.
8. Robaina-Estevez S., Daloso D. M., Zhang Y., Fernie A.R. & Nikoloski Z. (2017). Resolving the Central Metabolism of Arabidopsis Guard Cells. Scientific Reports 7(1): 8307.
7. Liu Z., Schneider R., Kesten C., Zhang Y., Somssich M., Zhang Y., Fernie A.R. & Persson S. (2016). Cellulose-Microtubule Uncoupling Proteins Prevent Lateral Displacement of Microtubules During Cellulose Synthesis in Arabidopsis. Developmental Cell 38(3): 305-315.
6. Liang C., Cheng S., Zhang Y., Sun Y., Fernie A.R., Kang K., Panagiotou G., Lo C. & Lim B.L. (2016). Transcriptomic, Proteomic and Metabolic Changes in Arabidopsis Thaliana Leaves after the Onset of Illumination. BMC Plant Biology 16: 43.
5. Pham P.A., Wahl V., Tohge T., de Souza L.R., Zhang Y., Do P.T., Olas J.J., Stitt M., Araujo W.L. & Fernie A.R. (2015). Analysis of Knockout Mutants Reveals Non-Redundant Functions of Poly(Adp-Ribose)Polymerase Isoforms in Arabidopsis. Plant Molecular Biology 89(4-5): 319-338.
4. Liang C., Zhang Y., Cheng S., Osorio S., Sun Y., Fernie A.R., Cheung C.Y. & Lim B.L. (2015). Impacts of High ATP Supply from Chloroplasts and Mitochondria on the Leaf Metabolism of Arabidopsis Thaliana. Frontiers in Plant Science 6: 922.
3. Zhang Y., Sun F., Fettke J., Schottler M.A., Ramsden L., Fernie A.R. & Lim B.L. (2014). Heterologous Expression of Atpap2 in Transgenic Potato Influences Carbon Metabolism and Tuber Development. FEBS Letters 588(20): 3726-3731.
2. Zhang Y., Yu L., Yung K.F., Leung D.Y.C., Sun F. & Lim B.L. (2012). Over-Expression of AtPAP2 in Camelina Sativa Leads to Faster Plant Growth and Higher Seed Yield. Biotechnology for Biofuels 5(1): 19.
1. Sun F., Suen P.K., Zhang Y., Liang C., Carrie C., Whelan J., Ward J.L., Hawkins N.D., Jiang L. & Lim B. L. (2012). A Dual-Targeted Purple Acid Phosphatase in Arabidopsis Thaliana Moderates Carbon Metabolism and Its Overexpression Leads to Faster Plant Growth and Higher Seed Yield. New Phytologist 194(1): 206-219.


 CAS
CAS
 中文
中文




.png)