Polycomb group (PcG) mediated H3K27me3 is commonly employed as a mechanism to repress developmentally important genes in both metazoans and plants. More than 7,000 genes, representing 21% of the genes in the Arabidopsis genome, are direct targets of H3K27me3. It is known that Jumonji C domain-containing proteins, namely KDM6a and KDM6b, are H3K27me3 demethylases in metazoan. Phylogenetic analysis did not reveal any orthologs of KDM6a or KDM6b in Arabidopsis or rice. Thus, although the existence of an active H3K27me3 demethylase activity is strongly implicated, how H3K27me3 is actively removed in plants remains unknown. Recent research in Xiaofeng Cao’s lab describes the long-sought-after H3K27me3 demethylase in Arabidopsis.
In this study, they used multiple approaches including genetics, biochemistry, molecular biology, transcriptomics and epigenomics to demonstrate that REF6 (RELATIVE OF EARLY FLOWERING 6), also known as JMJ12 (Jumonji domain-containing protein 12), functions as H3K27me3 demethylase. Through systematically screening for genes that reduce H3K27me3 level when overexpressed within potentially active JMJs, REF6 overexpression was found to remove H3K27me3/2 marks in vivo. In addition, immunoaffinity purified REF6-YFP-HA could demethylate oligonucleosomes at H3K27me3/2, but not H3K27me1, H3K4me3, H3K9me3 or H3K36me3 in vitro.Overexpression of REF6 in Arabidopsis leads to pleiotropic phenotypes similar to lhp1 and emf2 mutants that are defective in H3K27me3-mediated gene silencing. Moreover, consistent with the phenotypes, a dramatic reduction in H3K27me3/2 levels in REF6 overexpression lines was observed. This further demonstrated that REF6 acts as an H3K27me3/2 demethylase in vivo. The genetic interaction between ref6 and mutants of H3K27me3 methyltransferases clearly demonstrate that REF6 functions antagonistically with H3K27me3 methyltransferases. Finally, unbiased ChIP-seq epigenomic analysis revealed that hundreds of genes showed more than 3-fold hypermethylation in the ref6 loss-of-function mutant. In the ref6 mutant, H3K27me3 hypermethylated genes overlap with transcriptionally downregulated genes. Therefore, this work fills a major gap in understanding of the regulation of H3K27me3 in plants. It is worth to note that a fair number of genes regulated by REF6 are involved not only in developmental patterning as is known for most H3K27me3 targets, but also in many other processes such as response to internal and external stimuli. The genomics resources generated in this study will be valuable for future pursuits into various aspects of plant biology.
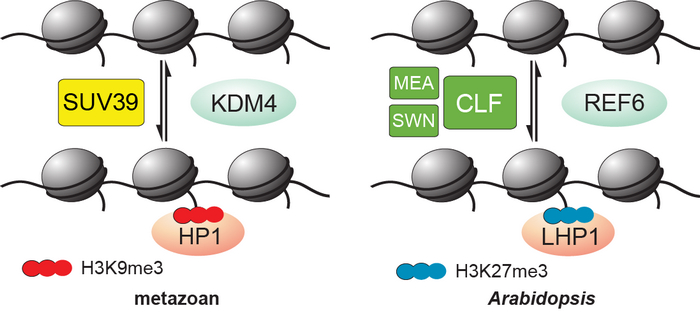
REF6 showed homology in sequence to metazoan H3K9me2/3 and H3K36me2/3 demethylases, KDM4 subfamily proteins. In metazoan H3K9me3 recruits HP1 to regulate gene expression. Arabidopsis genome harbors little H3K9me3 methylation. LHP1, the Arabidopsis homolog of HP1, binds to H3K27me3 rather than H3K9me3 to repress gene expression. This suggests that REF6 along with LHP1 may have acquired the novel function of acting in H3K27me3-mediated gene silencing during evolution. Before this study, the molecular basis of PcG depression is only understood in metazoan. Whether there is a uniform theme to counteract PcG remains a major question in the field of epigenetics. This work demonstrates that plants and metazoans employ conserved mechanisms in regulating H3K27me3 dynamics but use distinct subfamilies of enzymes.
This work entitled “Arabidopsis REF6 is a Histone H3 Lysine 27 Demethylase” was published online in Nature Genetics on June 5th, 2011. Graduate student Falong Lu and associated professor Xia Cui in Xiaofeng Cao’s lab are co-first authors of this work. This work was supported by the National Basic Research Program of China and by the National Natural Science Foundation of China.