The powdery mildew pathogens are biotrophic fungi that infect a large number of plant species, including major crops wheat and Barley, causing significant yield lossesworldwide.Previously, EDR2 (ENHANCED DISEASE RESISTANCE2) was identified as a critical regulator of powdery mildew resistance. The edr2 mutant displays enhanced resistance to powdery mildew and shows mildew-induced cell death phenotype. However, how EDR2 regulates defense responses is not well understood.
To study the molecular mechanisms of EDR2-mediated resistance, scientists in Dr. Dingzhong Tang’s lab from the Institute of Genetics and Developmental Biology, the Chinese Academy of Sciences examined the molecular interactions between Arabidopsis and the powdery mildew pathogen G. cichoracearum, by identifying and characterizing a suppressor of the edr2 mutant. They identified a mutation in RPN1a, which encodes a subunit of the 26S proteasome; this mutation suppresses all edr2-associated phenotypes, including powdery mildew resistance, mildew-induced cell death and ethylene-induced senescence. The rpn1a single mutant displayed enhanced susceptibility to virulent bacterial and fungal pathogens Pto DC3000 and G. cichoracearum, and to avirulent bacterial Pto DC3000 strains carrying avrRpt2 or avrRPS4. The rpn1a mutant has defects in salicylic acid (SA) accumulation in response to pathogen infection. Also, the accumulation of RPN1a protein is affected by SA. In addition, they showed that two other proteasome subunits, RPT2a and RPN8a are also involved in edr2-mediated disease resistance. However, mutations in several other proteasome subunits did not affect edr2-mediated resistance, suggesting that rpn1a-mediated enhanced susceptibility might not be caused by a general defect in 26S proteasome function. These findings provide evidence that some subunits of the 26S proteasome are directly involved in plant innate immunity inArabidopsis.
This work has recently been published online on The Plant Journal (DOI:10.1111/j.1365-313X.2012.05048.x), with Chunpeng Yao, a graduate student from Dr. Dingzhong Tang’s lab, as the first author. This research was supported by grants from National Basic Research Program of China, National Natural Science Foundation of China and National Transgenic Program of China.
AUTHOR CONTACT:
Dingzhong Tang, Ph.D.
Institute of Genetics and Developmetnal Biology, Chinese Academy of Sciences, Beijing, China.
E-mail: dztang@genetics.ac.cn
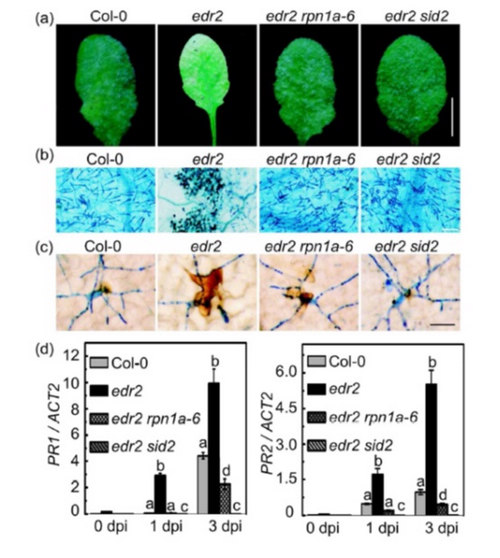
Figure. rpn1a-6 suppresses edr2-mediated resistance to G. cichoracearum. Five-week-old plants were infected with G. cichoracearum. (Image by Chunpeng Yao et al.)
(a) Leaves from the infected plants were photographed at 8 dpi. The edr2 mutant was resistant to G. cichoracearum, while the edr2 rpn1a-6 mutant displayed wild type-like susceptible phenotypes. Bar = 5mm
(b)Leaves were stained with trypan blue at 7 dpi. Note that large numbers of fungal spores were produced in wild type and edr2 rpn1a-6, but not in edr2. Bar = 100 μm
(c) Leaves were stained with 3, 3’-diaminobenzidine (DAB) and trypan blue sequentially at 2 dpi. Fungal structures are indicated by blue color, and hydrogen peroxide accumulation is indicated by brown color. Bar = 50 μm
(d) Quantitative RT-PCR analysis of PR1 and PR2 expression in the infected plants. The relative mRNA level was examined by Real-time PCR and normalized to ACTIN2. Bars represent mean and standard deviation (SD) of values obtained from three independent experiments. Different letters indicate statistically significant differences between samples (P < 0.05, Student’s t test).



