Small RNAs (smRNAs) in plants, mainly microRNAs and small interfering RNAs, play important roles in both transcriptional and post-transcriptional gene regulation. The broad application of high-throughput sequencing technology has made routinely generation of bulk smRNA sequences in laboratories possible, thus has significantly increased the need for batch analysis tools.
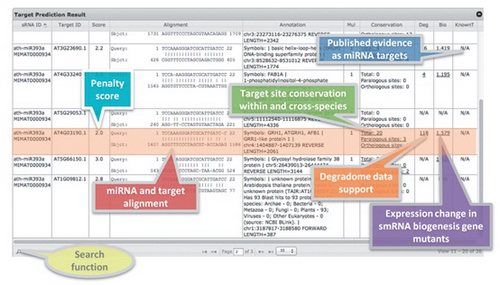
Scientists from Dr. Xiu-Jie Wang’s group of Institute of Genetics and Developmental Biology, the Chinese Academy of Sciences have developed an online smRNA analysis tool, named psRobot, which can identify smRNA with stem-loop shaped precursors (such as miRNA and short hairpin RNAs) and their target genes/transcripts requiring only smRNA sequences as input. psRobot has two modules: the first module is designed to identify stem-loop shaped smRNAs, including their expression in major plant smRNA biogenesis gene mutants and smRNA associated protein complexes to give clues to the smRNA generation and functional processes, their genome locations and precursor sequences. The second module is designed to return target prediction results of smRNAs, including smRNA target list, target multiplicity, target site conservation and biological data support, such as degradome data. Through these tools, users can easily acquire structure, biogenesis information and target genes of interesting smRNAs, which will largely facilitate the functional analysis of smRNAs in plants. PsRobot is freely accessible at http://omicslab.genetics.ac.cn/psRobot/.
The above work has been published online in Nucleic Acids Research (DOI:10.1093/nar/gks554) with Dr. Hua-Jun Wu and Ying-Ke Ma as the co-first authors. This study is supported by grants from National Natural Science Foundation of China, Chinese Academy of Sciences and Ministry of Agriculture of China.
AUTHOR CONTACT:
Xiu-Jie Wang, Ph.D., E-mail: xjwang@genetics.ac.cn
Meng Wang, Ph.D., E-mail: mengwang@genetics.ac.cn
(Image by Wu Hua-Jun et al.)
Figure 1. Result summary table of smRNA target prediction function.



