Gene regulatory networks (GRNs) control development via cell type-specific gene expression and interactions between transcription factors (TFs) and regulatory promoter regions. Plant organ boundaries separate lateral organs from the apical meristem and harbor axillary meristems (AMs). AMs, as stem cell niches, make the shoot a ramifying system. Although AMs have important functions in plant development, the knowledge of organ boundary and AM formation remains rudimentary.
In a recent study, researchers in Dr. JIAO Yuling’s group from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, generated a cellular resolution genome-wide gene expression map for low-abundance Arabidopsis thaliana organ boundary cells and constructed a genome-wide protein–DNA interaction map focusing on genes affecting boundary and AM formation. The resulting GRN uncovered transcriptional signatures, predicts cellular functions, and identified promoter hub regions that are bound by many TFs. Importantly, further experimental studies determined the regulatory effects of many TFs on their targets, identifying regulators and regulatory relationships in AM initiation. This systems biology approach thus enhanced people’s understanding of a key developmental process.
This work has been online published on the Molecular Systems Biology (http://onlinelibrary.wiley.com/doi/10.15252/msb.20145470/full), with research associate Dr. TIAN Caihuan as the first author.
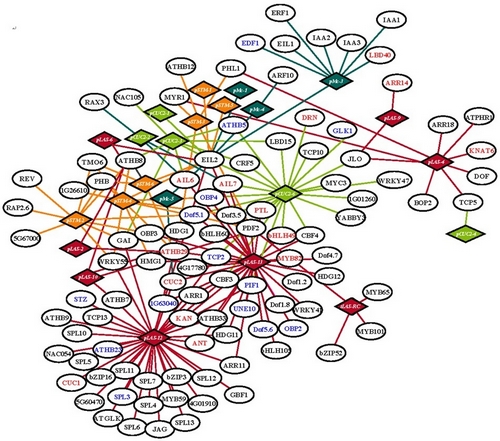
Figure1. A boundary-enriched protein-DNA interaction network. (Image by IGDB)
AUTHOR CONTACT:
JIAO Yuling, Ph.D.
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing, China.
Reference
Tian C, Zhang X, He J, Yu H, Wang Y, Shi B, Han Y, Wang G, Feng X, Zhang C, Wang J, Qi J, Yu R and Jiao Y. (2014). An organ boundary-enriched gene regulatory network uncovers regulatory hierarchies underlying axillary meristem initiation. Mol. Syst. Biol. 10: 755.



