In higher plants, meristems are responsible for the generation of all plant tissues and organs. While the shoot apical meristem (SAM) gives rise to all of the above-ground plant parts for the entire life of the plant through the continuous production of new organ primordial, the floral meristem (FM) will be terminated after the generation of all floral organs, known as FM determinacy, which helps ensure reproductive success, seed development, and, in applied cases, the yield of agricultural crops.
A research group led by Prof. LIU Xigang from the Center for Agricultural Resources Research (CARR), Institute of Genetics and Developmental Biology (IGDB) of Chinese Academy of Sciences (CAS) in collaboration with Prof. LIU Renyi from Shanghai Center for Plant Stress Biology, Shanghai Institutes for Biological Sciences (SIBS) of CAS discovered new function of FAR-RED ELONGATED HYPOCOTYL3 (FHY3) in plant meristem determinacy and maintenance by regulating SEPALLATA2 (SEP2) and CLAVATA3 (CLV3) expression.
It’s well known that FHY3 plays pivotal roles in the phyA signaling and the circadian clock pathways as well as other diverse developmental and physiological processes, including UV-B signaling, chloroplast biogenesis, chlorophyll biosynthesis, programmed cell death, ABA signaling, branching and flowering time at the plant vegetative stage. However, the roles of FHY3 in flower development remain unclear. In this study, researchers isolated several fhy3 mutations that enhanced the FM determinacy defects of ag-10, a weak agamous (ag) allele in a genetic screen. Genetic analysis showed that WUSCHEL (WUS) and CLV3, two central players in the establishment and maintenance of meristems, are epistatic to FHY3.
Using genome-wide ChIP-seq and RNA-seq data, researchers then identified hundreds of FHY3 target genes in flowers and find that FHY3 mainly acts as a transcriptional repressor in flower development, in contrast to its transcriptional activator role in seedlings. Binding motif-enrichment analyses indicated that FHY3 may co-regulate flower development with three flower specific MADS-domain TFs and four basic helix–loop–helix TFs that are involved in photomorphogenesis.
Further study revealed that CLV3, SEPALLATA1 (SEP1), and SEP2 are FHY3 target genes. In SAM, FHY3 directly represses CLV3, which consequently regulates WUS to maintain the stem cell pool. Intriguingly, CLV3 expression did not change significantly in fhy3 and phytochrome B mutants before and after light treatment, indicating that FHY3 and phytochrome B are involved in light-regulated meristem activity. In FM, FHY3 directly represses CLV3, but activates SEP2, to ultimately promote FM determinacy.
This work reveals new insights into the mechanisms of meristem maintenance and determinacy, and illustrates how the roles of a single TF may vary in different organs and developmental stages. More interestingly, FHY3 may act as a bridge molecule in the cross-talk between external signals and endogenous cues to coordinate plant development.
The article entitled “
FAR-RED ELONGATED HYPOCOTYL3 activates
SEPALLATA2 but inhibits
CLAVATA3 to regulate meristem determinacy and maintenance in
Arabidopsis” has been published online (
doi: 10.1073/pnas.1602960113) on July 28, 2016 in
Proceedings of the National Academy of Sciences of the United States of America.
This study was supported by National Science Foundation of China (NSFC), National Basic Research Program of China.
Figure1. FHY3 is required for floral meristem determinacy and shoot apical meristem maintenance. (Reprinted from Li et al., 2016)
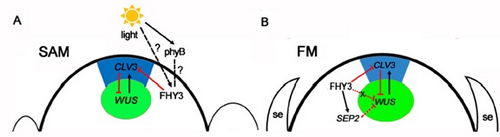
Figure2. Proposed model of FHY3 functions in meristem activity regulation. (Reprinted from Li et al., 2016)
Contact:
Dr. LIU Xigang
Center for Agricultural Resources Research, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences



