Soybean is an important crop for oil and protein resources. Its yield is contributed by many factors and 100-seed weight is one of these. Until now, 94 QTLs for seed weight have been identified (
http://www.soybase.org/ ) in soybean. However, the relevant genes and the mechanisms involved in seed weight control are still less understood.
By analysis of RILs derived from a cross between Glycine soja ZYD7 and Glycine max HN44, Prof ZHANG Jinsong’s group and Prof CHEN Shouyi’s group, from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, in collaboration with Prof Man Weiqun’s group and Prof LAI Yongcai’s group from Heilongjing Academy of Agricultural Sciences, identified a line R245 with higher 100-seed weight than that of HN44.
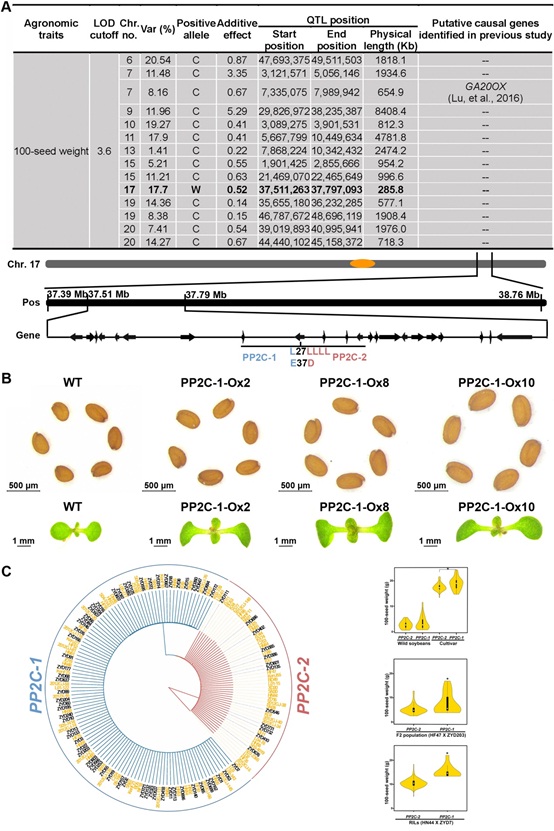
After whole-genome re-sequencing of 198 RILs and two parents, they acquired SNP data with high accuracy and quality, and constructed a genetic map. Based on this map, the genetic loci controlling the 100-seed weight and oil content of soybean were identified. The line R245 was found to contain 13 positive alleles from HN44 and 1 positive allele from ZYD7 after genotyping (Fig 1A). In the 285 kb region of the one positive allele from ZYD7, 22 protein-coding genes were predicted and none of these genes showed expression difference between ZYD7 and HN44 at the stage of early- and mid-maturation of soybean. However, two (Glyma17g33690: PP2C, Glyma17g33790: EAL) out of 22 genes showed nucleotide variation which resulted in amino acid changes in the encoded proteins.
By transgenic analysis in Arabidopsis, PP2C-1 from ZYD7 enhanced seed size/weight of the transgenic plant, whereas the PP2C-2 from HN44, EAL-1 from ZYD7 and EAL-2 from HN44 did not have this function (Fig 1B). By luciferase complementary imaging analysis, we found that PP2C-1 can interact with AtBZR1, AtBES1, GmBZR1, GmBZR2, GmBZR3 and GmBZR4, whereas PP2C-2 did not show interactions with these proteins. Further, PP2C-1 can dephosphorylate GmBZR1. In the PP2C-1-overexpressing transgenic plant, 11 genes relevant to seed size were expressed at higher levels compared to wild type Col-0. These results indicate that PP2C-1 may interact with the component of BR pathway to activate downstream genes for seed size/weight promotion.
By phylogenetic analysis, we found that a significant portion of the soybean cultivars did not contain the PP2C-1 allele (Fig 1C), and these cultivars may be improved for seed size/weight by introducing the PP2C-1 allele.
This work has been published online in
Mol Plant., on Mar 28, 2017 (
doi:10.1016/j.molp.2017.03.006). This work is supported by Strategic Priority Research Program of CAS and National Transgenic Research Projects etc.
 |
|
Fig 1. Identification of 100-seed weight genes from soybean (Image by IGDB). |
|
(A), Mapping of QTLs related to 100-seed weight using resequencing data from a RIL population. C: the positive allele comes from cultivar HN44; W: the positive allele comes from wild soybean ZYD7. PP2C-1 has an L at 27thposition and an E at 37th. PP2C-2 has four L and a D at the corresponding positions, respectively.
(B), Overexpression of PP2C-1 in transgenic Arabidopsis plants increases seed size/weight and cotyledon size.
(C), Distribution of PP2C-1 in soybean accessions. Left: ~40% of soybean accessions do not have PP2C-1 allele; Names in black indicate wild soybeans, and names in yellow indicate cultivars. Right upper: Association analysis of 100-seed weight with gene haplotypes in wild and cultivated soybeans. Right middle: Association analysis of 100-seed weight with gene haplotypes in F2 populations from ZYD203 and HF47. Right lower: Association analysis of 100-seed weight with gene haplotypes in RILs from ZYD7 and HN44. |



